This post includes the daily entry notebook for Sept-Oct 2022 Acropora coral spawning and field expedition in Moorea, French Polynesia. These notebook posts are updated in order of most recent to oldest.
Project Overview
The goal of this field expedition is to conduct spawning research in September and October in Acropora spp. Further, we will be continuing the monthly gametogenesis timeseries, sampling Acropora pulchra and Pocillopora spp. throughout gametogenesis on the northshore backreef to collect data on histology, physiology, and epigenomics. Original tagging and sampling of 40 Acropora pulchra colonies was completed by the Putnam Lab team (Hollie, Ariana, Pierrick, and Alex (CRIOBE)) in December 2021 and of Pocillopora spp. starting in January 2022. Monthly timepoint sampling will be carried out for this project until October 2022 for spawning. We will be processing physiological measurements (symbiont densities, chlorophyll content, total protein, surface area, and tissue biomass) for collected and preserved samples, measuring bleaching score on in situ colonies, decalcifying histological samples, and collecting molecular and histological samples during our three month trip.
A. Huffmyer is conducting field work to investigate metabolic integration of coral host and symbiont in early life history as part of funded National Science Foundation Ocean Sciences Postdoctoral Fellowship and the University of Washington eScience Data Science Postdoctoral Fellowship. More information on the abstract of this award can be found here. Ariana’s notebook can be viewed here.
D. Becker Polinksi is conducting field work to characterize maternal provisioning, epigenetic inheritance, and cross-generational impacts of thermal stress on Acropora pulchra throughout gametogenesis and early development (i.e., embryo, larval development, recruits, and juveniles) with parents that have previously experienced a marine heatwave event in March-April 2022. This research will connect molecular mechanisms impacts on phenotypic response through gametogenesis, larval development, and post settlement, to help understand acclimatization patterns facilitated by parental inheritance. All data and further information can be found on her personal GitHub here.
We will also continue in situ monitoring of Acropora and Pocillopora colonies on the reef in Moorea.
Finally, we will conduct an experiment to expose adult Acropora pulchra corals to conditions that elicit changes in symbiotic and metabolic condition and test how these changes influence response to a repeat stressor. We are manipulating light and feeding conditions to elicit metabolic and symbiotic responses and testing stress response at the end of the experiment using the thermal stress CBASS system. Notebook post detailing activities specific to the E5 adult metabolism and stressor experiment (details below) can be found here and on the E5 A. pulchar Metabolism GitHub repository.
Field expedition members:
Ariana Huffmyer
Pierrick Harnay
Danielle Becker-Polinski
Hollie Putnam
28 October 2022
Huffmyer - Final sampling - day 5 of experiment
Today is the final day of sampling in our experiment! Notebook posts regarding future data analyses will be found in Ariana’s GitHub Notebook.
This sampling will be on day 5 of the experiment after the larvae have experienced 3.5 days of temperature stress (temperature started Monday PM). We have been dosing symbionts into the tank systems for 72 hours (Tues, Wed, and Thurs). As a reminder, these are our treatments:
| Tank | Symbiont Treatment | Temperature Treatment |
|---|---|---|
| A1 | Added symbionts | High (+2-2.5°C) |
| A2 | Added symbionts | High (+2-2.5°C) |
| A3 | Added symbionts | High (+2-2.5°C) |
| A4 | No symbionts | High (+2-2.5°C) |
| A5 | No symbionts | High (+2-2.5°C) |
| A6 | No symbionts | High (+2-2.5°C) |
| B1 | Added symbionts | Ambient |
| B2 | Added symbionts | Ambient |
| B3 | Added symbionts | Ambient |
| B4 | No symbionts | Ambient |
| B5 | No symbionts | Ambient |
| B6 | No symbionts | Ambient |
Sampling protocols followed those described from 24 October 2022.
- 1. Obtain larvae: Larvae were collected from each tank by pouring all water from each tank through a seive to collect all larvae. Larvae were added to labeled falcon tubes and brought in right before that tank needed to be sampled. We first brought in larvae for respirometry measurements. At the time of taking larvae out of tanks we measured larval density to estimate the number of larvae that we had for the day.
| Tank | Time | Pool Volume | Rep 1 | Rep 2 | Rep 3 | Rep 4 | Rep 5 | Rep 6 | Rep Volume mL | Larvae per mL | Total Larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 645 | 310 | 1 | 3 | 2 | 0 | 1 | 3 | 1 | 1.6 | 500 |
| A2 | 325 | 4 | 2 | 2 | 4 | 4 | 2 | 1 | 3 | 975 | |
| A3 | 375 | 1 | 1 | 3 | 3 | 2 | 2 | 1 | 2 | 750 | |
| A4 | 400 | 1 | 2 | 2 | 5 | 1 | 2 | 1 | 2.16 | 866 | |
| A5 | 300 | 4 | 1 | 4 | 3 | 2 | 3 | 1 | 2.8 | 850 | |
| A6 | 300 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 1 | 300 | |
| B1 | 400 | 2 | 1 | 2 | 1 | 1 | 0 | 1 | 1.16 | 366 | |
| B2 | 175 | 2 | 3 | 4 | 3 | 1 | 1 | 1 | 2.3 | 408 | |
| B3 | 280 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 1.16 | 326 | |
| B4 | 400 | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 0.83 | 333 | |
| B5 | 350 | 5 | 1 | 1 | 3 | 4 | 1 | 1 | 2.5 | 875 | |
| B6 | 745 | 300 | 2 | 3 | 2 | 4 | 2 | 4 | 1 | 2.83 | 850 |
We have limited larvae especially in tanks A6, B1, B3, and B4. We will look at symbiont cell density next to decide if we have enough larvae with symbionts to do stable isotope incubations. We will have enough to do the standard sampling that we have done the last two time points.
The larvae were divided into two falcon tubes so that one at a time could be brought into the lab for counting and the other can stay in the treatment tank. The falcon tubes also contained some algae and debris from emptying the entire tank, so this gave more water to each falcon tube so that the larvae weren’t overly stocked.
-
2. Cell counts: We first sampled n=8 larvae from each tank at 0800 during the pooling process to look at cell densities. There were 0 cells in all larvae that we looked at. Because of this, we will not do stable isotope incubations today. We will sample for respirometry, metabolomics, and RNA/DNA as we have done on 24 and 26 October 2022. This will provide data on a time series of metabolism in larvae under elevated temperature prior to symbiont uptake.
-
3. Respirometry: We ran a total of 2 runs of respirometry (4 total plates) today. We did not run the additional 2 runs that we did on 26 October 2022 because we had limited larvae to work with. The plate maps and information for each run are below. Protocols followed 24 October 2022. Plate maps, temperature, and light data were recorded on Google Drive. All wells have 5 larvae unless noted below.
Plate 13; SDR 1; SDR 641; 27°C; 20221028
Time start: 9:15
Time R start: 9:35
Time end: 9:50
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A1 | A2 | A3 | B1 | B2 | B3 |
| B | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| C | A1 | A2 | A3 | B1 | B2 | B3 |
| D | A1 | A2 | A3 | B1 | B2 | B3 |
Well D6: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 915 | 27.72 |
| 920 | 27.53 |
| 930 | 28.72 |
| 940 | 28.7 |
| Light measurements | |
|---|---|
| Position | Light |
| Center | 519 |
| Q1 | 507 |
| Q2 | 488 |
| Q3 | 471 |
| Q4 | 506 |
Plate 14; SDR 2; SDR 710; 30°C; 20221028
Time start: 9:15
Time R start: 9:35
Time end: 9:50
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A1 | A2 | A3 | B1 | B2 | B3 |
| B | A1 | A2 | A3 | B1 | B2 | B3 |
| C | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| D | A1 | A2 | A3 | B1 | B2 | B3 |
Well B1: 4 larvae Well D4: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 915 | 30.71 |
| 920 | 30.89 |
| 930 | 31.42 |
| 938 | 31.62 |
| Light measurements | |
|---|---|
| Position | Light |
| Center | 484 |
| Q1 | 517 |
| Q2 | 488 |
| Q3 | 485 |
| Q4 | 505 |
Plate 15; SDR 1; SDR 641; 27°C; 20221028
Time start: 10:35
Time R start: 10:55
Time end: 11:10
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A4 | A5 | A6 | B4 | B5 | B6 |
| B | A4 | A5 | A6 | B4 | B5 | B6 |
| C | A4 | A5 | A6 | B4 | B5 | B6 |
| D | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
Well B4: 4 larvae
Well B5: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1033 | 28.07 |
| 1040 | 28.83 |
| 1050 | 28.24 |
| 1102 | 27.97 |
Plate 16; SDR 2; SDR 710; 30°C; 20221028
Time start: 10:35
Time R start: 10:55
Time end: 11:10
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A4 | A5 | A6 | B4 | B5 | B6 |
| B | A4 | A5 | A6 | B4 | B5 | B6 |
| C | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| D | A4 | A5 | A6 | B4 | B5 | B6 |
Well D5: 4 larvae
Well D4: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1033 | 32.14 |
| 1040 | 30.97 |
| 1050 | 29.33 |
| 1102 | 30.92 |
- 4. Sampling for metabolomics and RNA/DNA: Larvae were sampled today using the same protocol as on 24 October 2022. Larvae were counted from tanks in random order and placed in an incubator at the treatment temperature (either 27°C or 30°C) for 10-15 minutes prior to sampling. Larvae in falcon tubes were kept in the treatment tanks (i.e., a water bath) until counted.
The tubes sampled today were:
R73-R108 (n=3 tubes per tank, n=50 larvae per tube)
M73-M108 (n=3 tubes per tank, n=50 larvae per tube)
| Tube ID | Date | Time | Tank | Number larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| R73 | 20221028 | 1346 | A1 | 50 | RNA DNA Shield 600uL | -40C |
| R74 | 20221028 | 1346 | A1 | 50 | RNA DNA Shield 600uL | -40C |
| R75 | 20221028 | 1346 | A1 | 50 | RNA DNA Shield 600uL | -40C |
| R76 | 20221028 | 1410 | A2 | 50 | RNA DNA Shield 600uL | -40C |
| R77 | 20221028 | 1410 | A2 | 50 | RNA DNA Shield 600uL | -40C |
| R78 | 20221028 | 1410 | A2 | 50 | RNA DNA Shield 600uL | -40C |
| R79 | 20221028 | 1539 | A3 | 50 | RNA DNA Shield 600uL | -40C |
| R80 | 20221028 | 1539 | A3 | 50 | RNA DNA Shield 600uL | -40C |
| R81 | 20221028 | 1539 | A3 | 50 | RNA DNA Shield 600uL | -40C |
| R82 | 20221028 | 1440 | A4 | 50 | RNA DNA Shield 600uL | -40C |
| R83 | 20221028 | 1440 | A4 | 50 | RNA DNA Shield 600uL | -40C |
| R84 | 20221028 | 1440 | A4 | 50 | RNA DNA Shield 600uL | -40C |
| R85 | 20221028 | 1208 | A5 | 50 | RNA DNA Shield 600uL | -40C |
| R86 | 20221028 | 1208 | A5 | 50 | RNA DNA Shield 600uL | -40C |
| R87 | 20221028 | 1208 | A5 | 50 | RNA DNA Shield 600uL | -40C |
| R88 | 20221028 | 1505 | A6 | 50 | RNA DNA Shield 600uL | -40C |
| R89 | 20221028 | 1505 | A6 | 50 | RNA DNA Shield 600uL | -40C |
| R90 | 20221028 | 1505 | A6 | 50 | RNA DNA Shield 600uL | -40C |
| R91 | 20221028 | 1500 | B1 | 50 | RNA DNA Shield 600uL | -40C |
| R92 | 20221028 | 1500 | B1 | 50 | RNA DNA Shield 600uL | -40C |
| R93 | 20221028 | 1500 | B1 | 50 | RNA DNA Shield 600uL | -40C |
| R94 | 20221028 | 1555 | B2 | 50 | RNA DNA Shield 600uL | -40C |
| R95 | 20221028 | 1555 | B2 | 50 | RNA DNA Shield 600uL | -40C |
| R96 | 20221028 | 1555 | B2 | 50 | RNA DNA Shield 600uL | -40C |
| R97 | 20221028 | 1450 | B3 | 50 | RNA DNA Shield 600uL | -40C |
| R98 | 20221028 | 1450 | B3 | 50 | RNA DNA Shield 600uL | -40C |
| R99 | 20221028 | 1450 | B3 | 50 | RNA DNA Shield 600uL | -40C |
| R100 | 20221028 | 1426 | B4 | 50 | RNA DNA Shield 600uL | -40C |
| R101 | 20221028 | 1426 | B4 | 50 | RNA DNA Shield 600uL | -40C |
| R102 | 20221028 | 1426 | B4 | 50 | RNA DNA Shield 600uL | -40C |
| R103 | 20221028 | 1215 | B5 | 50 | RNA DNA Shield 600uL | -40C |
| R104 | 20221028 | 1215 | B5 | 50 | RNA DNA Shield 600uL | -40C |
| R105 | 20221028 | 1215 | B5 | 50 | RNA DNA Shield 600uL | -40C |
| R106 | 20221028 | 1535 | B6 | 50 | RNA DNA Shield 600uL | -40C |
| R107 | 20221028 | 1535 | B6 | 50 | RNA DNA Shield 600uL | -40C |
| R108 | 20221028 | 1535 | B6 | 50 | RNA DNA Shield 600uL | -40C |
| Tube ID | Date | Time | Tank | Number larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| M73 | 20221028 | 1346 | A1 | 50 | Flash frozen | -40C |
| M74 | 20221028 | 1346 | A1 | 50 | Flash frozen | -40C |
| M75 | 20221028 | 1346 | A1 | 50 | Flash frozen | -40C |
| M76 | 20221028 | 1410 | A2 | 50 | Flash frozen | -40C |
| M77 | 20221028 | 1410 | A2 | 50 | Flash frozen | -40C |
| M78 | 20221028 | 1410 | A2 | 50 | Flash frozen | -40C |
| M79 | 20221028 | 1539 | A3 | 50 | Flash frozen | -40C |
| M80 | 20221028 | 1539 | A3 | 50 | Flash frozen | -40C |
| M81 | 20221028 | 1539 | A3 | 50 | Flash frozen | -40C |
| M82 | 20221028 | 1440 | A4 | 50 | Flash frozen | -40C |
| M83 | 20221028 | 1440 | A4 | 50 | Flash frozen | -40C |
| M84 | 20221028 | 1440 | A4 | 50 | Flash frozen | -40C |
| M85 | 20221028 | 1208 | A5 | 50 | Flash frozen | -40C |
| M86 | 20221028 | 1208 | A5 | 50 | Flash frozen | -40C |
| M87 | 20221028 | 1208 | A5 | 50 | Flash frozen | -40C |
| M88 | 20221028 | 1505 | A6 | 50 | Flash frozen | -40C |
| M89 | 20221028 | 1505 | A6 | 50 | Flash frozen | -40C |
| M90 | 20221028 | 1505 | A6 | 50 | Flash frozen | -40C |
| M91 | 20221028 | 1500 | B1 | 50 | Flash frozen | -40C |
| M92 | 20221028 | 1500 | B1 | 50 | Flash frozen | -40C |
| M93 | 20221028 | 1500 | B1 | 50 | Flash frozen | -40C |
| M94 | 20221028 | 1555 | B2 | 50 | Flash frozen | -40C |
| M95 | 20221028 | 1555 | B2 | 50 | Flash frozen | -40C |
| M96 | 20221028 | 1555 | B2 | 50 | Flash frozen | -40C |
| M97 | 20221028 | 1450 | B3 | 50 | Flash frozen | -40C |
| M98 | 20221028 | 1450 | B3 | 50 | Flash frozen | -40C |
| M99 | 20221028 | 1450 | B3 | 50 | Flash frozen | -40C |
| M100 | 20221028 | 1426 | B4 | 50 | Flash frozen | -40C |
| M101 | 20221028 | 1426 | B4 | 50 | Flash frozen | -40C |
| M102 | 20221028 | 1426 | B4 | 50 | Flash frozen | -40C |
| M103 | 20221028 | 1215 | B5 | 50 | Flash frozen | -40C |
| M104 | 20221028 | 1215 | B5 | 50 | Flash frozen | -40C |
| M105 | 20221028 | 1215 | B5 | 50 | Flash frozen | -40C |
| M106 | 20221028 | 1535 | B6 | 50 | Flash frozen | -40C |
| M107 | 20221028 | 1535 | B6 | 50 | Flash frozen | -40C |
| M108 | 20221028 | 1535 | B6 | 50 | Flash frozen | -40C |
After sampling, there were not enough larvae left to sample for physiological metrics. This is the end of larval sampling!
Larval samples are stored at -40°C in the molecular freezer at Gump until transport to the US (approved by CITES) in December 2022.
Huffmyer - Downloading loggers
All Hobo pendant loggers were downloaded from larval tanks at 1600.
Huffmyer - Cleaning tanks
All tanks were cleaned at 1700-1800.
The experiment is complete!
27 October 2022
Huffmyer - Symbiont innoculation
Symbionts were isolated from 6 parent fragments following protocols from 24 October 2022. The calculations were as follows:
| Date | Time | Total Volume mL | Squares Counted | Count1 | Count2 | Count3 | Count4 | Count5 | Count6 | Mean Count | Cells per mL | Cells total | Notes | Date | Pool | Symbiont Density (C1) | Target Density (C2) | Volume Symbionts (V1) | Tank volume (V2) | Time Added/Water Off | Time Water On | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20221027 | 745 | 45 | 1 | 200 | 190 | 185 | 172 | 186.75 | 18675000 | 840375000 | 20221027 | 5 parents | 840375000 | 30000 | 0.6425702811 | 18000 | 745 | 845 | ||||
| 20221027 | 1540 | 45 | 1 | 200 | 190 | 185 | 172 | 186.75 | 18675000 | 840375000 | 20221027 | 5 parents | 840375000 | 30000 | 0.6425702811 | 18000 | 1540 | 1640 |
0.9 mL of symbiont isolate was added to each symbiont addition tank (A1-3, B1-3) at 0745 with water turned off until 0845.
The remaining symbiont isolate was stored at 4°C until the evening dosing. The evening dosing was added at 1540 with water turned off until 1640 with 0.9 mL per tank.
Returning colonies
We returned all parent colonies to the Mahana field site from 0800-1100 with Hollie and Danielle and Tony.
Huffmyer - Taking extra samples
Ariana and Terava collected extra larval samples to optimize and practice extraction protocols without sacrificing experimental samples. These were sampled using the same methods as described on 24 October 2022. The following samples were collected across a gradient of the number of larvae from 1-100. These larvae were taken from the 20 October 2022 cohort that had been reared in coolers since spawning with water changes every 2 days by the Correa Lab.
| Tube ID | Date | Time | Tank | Number larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| M109 | 20221027 | 1509 | Extra | 1 | Flash frozen | -40C |
| M110 | 20221027 | 1509 | Extra | 1 | Flash frozen | -40C |
| M111 | 20221027 | 1509 | Extra | 1 | Flash frozen | -40C |
| M112 | 20221027 | 1512 | Extra | 5 | Flash frozen | -40C |
| M113 | 20221027 | 1512 | Extra | 5 | Flash frozen | -40C |
| M114 | 20221027 | 1512 | Extra | 5 | Flash frozen | -40C |
| M115 | 20221027 | 1520 | Extra | 10 | Flash frozen | -40C |
| M116 | 20221027 | 1520 | Extra | 10 | Flash frozen | -40C |
| M117 | 20221027 | 1520 | Extra | 10 | Flash frozen | -40C |
| M118 | 20221027 | 1530 | Extra | 30 | Flash frozen | -40C |
| M119 | 20221027 | 1530 | Extra | 30 | Flash frozen | -40C |
| M120 | 20221027 | 1530 | Extra | 30 | Flash frozen | -40C |
| M121 | 20221027 | 1540 | Extra | 50 | Flash frozen | -40C |
| M122 | 20221027 | 1540 | Extra | 50 | Flash frozen | -40C |
| M123 | 20221027 | 1540 | Extra | 50 | Flash frozen | -40C |
| M124 | 20221027 | 1550 | Extra | 100 | Flash frozen | -40C |
| M125 | 20221027 | 1550 | Extra | 100 | Flash frozen | -40C |
| M126 | 20221027 | 1550 | Extra | 100 | Flash frozen | -40C |
| Tube ID | Date | Time | Tank | Number larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| R109 | 20221027 | 1345 | Extra | 1 | RNA DNA Shield 600uL | -40C |
| R110 | 20221027 | 1345 | Extra | 1 | RNA DNA Shield 600uL | -40C |
| R111 | 20221027 | 1345 | Extra | 1 | RNA DNA Shield 600uL | -40C |
| R112 | 20221027 | 1348 | Extra | 5 | RNA DNA Shield 600uL | -40C |
| R113 | 20221027 | 1348 | Extra | 5 | RNA DNA Shield 600uL | -40C |
| R114 | 20221027 | 1348 | Extra | 5 | RNA DNA Shield 600uL | -40C |
| R115 | 20221027 | 1358 | Extra | 10 | RNA DNA Shield 600uL | -40C |
| R116 | 20221027 | 1358 | Extra | 10 | RNA DNA Shield 600uL | -40C |
| R117 | 20221027 | 1358 | Extra | 10 | RNA DNA Shield 600uL | -40C |
| R118 | 20221027 | 1402 | Extra | 30 | RNA DNA Shield 600uL | -40C |
| R119 | 20221027 | 1402 | Extra | 30 | RNA DNA Shield 600uL | -40C |
| R120 | 20221027 | 1402 | Extra | 30 | RNA DNA Shield 600uL | -40C |
| R121 | 20221027 | 1420 | Extra | 50 | RNA DNA Shield 600uL | -40C |
| R122 | 20221027 | 1420 | Extra | 50 | RNA DNA Shield 600uL | -40C |
| R123 | 20221027 | 1420 | Extra | 50 | RNA DNA Shield 600uL | -40C |
| R124 | 20221027 | 1445 | Extra | 100 | RNA DNA Shield 600uL | -40C |
| R125 | 20221027 | 1445 | Extra | 100 | RNA DNA Shield 600uL | -40C |
| R126 | 20221027 | 1445 | Extra | 100 | RNA DNA Shield 600uL | -40C |
We also collected extra samples for physiology analyses. We can use these to test physiology assays that we may collect for in future projects.
| Tube ID | Date | Time | Tank | Volume larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| P37 | 20221027 | 1625 | Extra | 50 larvae | Flash freeze | -40C |
| P38 | 20221027 | 1625 | Extra | 50 larvae | Flash freeze | -40C |
| P39 | 20221027 | 1625 | Extra | 50 larvae | Flash freeze | -40C |
| P40 | 20221027 | 1625 | Extra | 50 larvae | Flash freeze | -40C |
| P41 | 20221027 | 1625 | Extra | 50 larvae | Flash freeze | -40C |
| P42 | 20221027 | 1625 | Extra | 50 larvae | Flash freeze | -40C |
Terava assisted with this sample collection today.

Huffmyer - Preparing isotope labeling and solutions
Ariana prepared calculations and labels for stable isotope incubations in case we decide to move forward with this tomorrow. We will collect cell density samples tomorrow morning and if the larvae have symbionts, we will do the stable isotope incubations. If the larvae do not have symbionts, we will not do stable isotope metabolomics since this method is meant to detect nutritional exchange between host and symbiont. We will use Ariana’s larval stable isotope protocol.
In the case that we decide to move forward, these are the replicates we will need. Note taht we are only going to do incubations in the symbiotic treatments if symbionts are present (tanks A1, 2, and 3 and tanks B1, 2, and 3).
- n=6 vials of 50 larvae from each tank incubated with C13 label at their respective treatment temperature (B tanks at 27°C; A tanks at 30°C); N=36 vials total
- n=2 vials of 50 larvae from a pool of each temperature treatment (pool of A and pool of B tanks) incubated with C12 sodium bicarbonate (no label) each at 27°C and 30°C; N=4 vials total
- n=2 vials of 50 larvae from a pool of each temperature treatment (pool of A and pool of B tanks) incubated with C13 label each at 27°C and 30°C in the dark (covered with aluminum foil); N=4 vials total
- n=3 vials of 15 larvae from a pool of each temperature treatment (pool of A and pool of B tanks) incubated with C13 label each at 27°C and 30°C that will be preserved for trial MALDI microscopy with J. Matthews at UT Sydney; N=6 vials total
- n=3 vials of 50 larvae from a pool of each temperature treatment (pool of A and pool of B tanks) incubated with C13 label each at 27°C and 30°C that will be preserved for trial lipid-protein-metabolite extractions with J. Matthews at UT Sydney; N=6 vials total
Total vials with C13 = 36 + 4 + 6 + 6 = 52 C13 vials Total vials with C12 = 4
| Number of vials | |
|---|---|
| 27C | 30 C |
| 6 of A1 | 6 of B1 |
| 6 of A2 | 6 of B2 |
| 6 of A3 | 6 of B3 |
| 2 C12 | 2 C12 |
| 2 Dark C13 | 2 Dark C13 |
| 3 MALDI | 3 MALDI |
| 3 Tri Extract | 3 Tri Extract |
The solutions we will need are calculated below.
| Solution | Vials | Falcon tubes | Total volume needed (mL) | FSW | C13 (mg) | C12 (mg) | Concentration |
|---|---|---|---|---|---|---|---|
| C13 | 52 | 0 | 1040 | 1200 | 462.8571429 | 0 | 4 mM solution |
| C12 | 4 | 0 | 80 | 100 | 0 | 41.66666667 | 4 mM solution |
The solutions will be made by adding the required C13 or C12 to 0.02 µm filtered seawater with the water brought to treatment temperature before addition to the vials. Vials will then be incubated under the light at 200 PAR for 4-5 hours and sampled as described in the larval stable isotope protocol.
If needed we will also make a 3% sodium abscorbate solution to preserve the MALDI samples. The samples (n=15 larave each) will be submerged in the sodium abscorbate solution and then flash frozen in liquid nitrogen as required for this microscopy analysis. To make the solution we will add 0.15 g of sodium abscorbate to 5 mL of 0.02 µm FSW.
We will decide in the morning if we will move forward with the stable isotope work. Vials were prelabeled with tank ID in case we decide to use them.
Larval rearing
Daily measurements were taken at 1800-1830 by Ariana. It was already dark, so light measurements were not taken.
Larvae were stirred and maintained, temperature treatments are on track, and filters were cleaned every 4-5 hours.
26 October 2022
Huffmyer - Sampling - day 3 of experiment
All sampling protocols follow what we did on 24 October 2022. Details of sampling are included below if they deviate from the standard protocol on 24 October 2022.
- 1. Obtain larvae: Larvae were collected from each tank by pouring water through a seive until the number of larvae needed was obtained (~350-400). Larvae were added to labelled falcon tubes and brought in right before that tank needed to be sampled. We first brought in larvae for respirometry measurements.
- 2. Respirometry: We ran a total of 4 runs of respirometry (8 total plates) today. The plate maps and information for each run are below. Protocols followed 24 October 2022. Plate maps, temperature, and light data were recorded on Google Drive. All wells have 5 larvae.
Plate 5: SDR 1; SDR 641; 27°C; 20221026
Time start: 9:26
Time R start: 9:46
Time end: 10:04
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| B | A4 | A5 | A6 | B4 | B5 | B6 |
| C | A4 | A5 | A6 | B4 | B5 | B6 |
| D | A4 | A5 | A6 | B4 | B5 | B6 |
Well D1: 4 larvae
Well B3: 3 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 943 | 28.03 |
| 950 | 27.95 |
| 958 | 27.88 |
| Light measurements | |
|---|---|
| Position | Light |
| Center | 538 |
| Q1 | 610 |
| Q2 | 533 |
| Q3 | 487 |
| Q4 | 485 |
Plate 6: SDR 2; SDR 710; 30°C; 20221026
Time start: 9:26
Time R start: 9:46
Time end: 10:04
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A4 | A5 | A6 | B4 | B5 | B6 |
| B | A4 | A5 | A6 | B4 | B5 | B6 |
| C | A4 | A5 | A6 | B4 | B5 | B6 |
| D | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
Well C6: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 943 | 30.22 |
| 950 | 30.45 |
| 958 | 31.03 |
| Light measurements | |
|---|---|
| Position | Light |
| Center | 512 |
| Q1 | 506 |
| Q2 | 510 |
| Q3 | 462 |
| Q4 | 447 |
Plate 7: SDR 1; SDR 641; 27°C; 20221026
Time start: 10:33
Time R start: 10:53
Time end: 11:08
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A1 | A2 | A3 | B1 | B2 | B3 |
| B | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| C | A1 | A2 | A3 | B1 | B2 | B3 |
| D | A1 | A2 | A3 | B1 | B2 | B3 |
Well D4: 4 larvae
Well D1: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1033 | 27.66 |
| 1043 | 27.53 |
| 1051 | 28.12 |
| 1055 | 27 |
| 1102 | 28.55 |
Plate 8: SDR 2; SDR 710; 30°C; 20221026
Time start: 10:33
Time R start: 10:53
Time end: 11:08
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A1 | A2 | A3 | B1 | B2 | B3 |
| B | A1 | A2 | A3 | B1 | B2 | B3 |
| C | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| D | A1 | A2 | A3 | B1 | B2 | B3 |
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1033 | 31.04 |
| 1045 | 31.4 |
| 1051 | 31.42 |
| 1055 | 26.91 |
| 1102 | 30.11 |
Plate 9: SDR 1; SDR 641; 27°C; 20221026
Time start: 11:32
Time R start: 11:49
Time end: 12:04
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| B | B4 | B5 | B6 | A4 | A5 | A6 |
| C | B4 | B5 | B6 | A4 | A5 | A6 |
| D | B4 | B5 | B6 | A4 | A5 | A6 |
Well D4: 4 larvae Well C4: 4 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1137 | 28.88 |
| 1141 | 28.53 |
| 1145 | 28.31 |
| 1149 | 26.51 |
| 1159 | 27.4 |
Plate 10: SDR 2; SDR 710; 30°C; 20221026
Time start: 11:32
Time R start: 11:49
Time end: 12:04
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | B4 | B5 | B6 | A4 | A5 | A6 |
| B | B4 | B5 | B6 | A4 | A5 | A6 |
| C | B4 | B5 | B6 | A4 | A5 | A6 |
| D | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
Well C2: 4 larvae
Well B3: 4 larvae
Well B5: 3 larvae
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1137 | 30.66 |
| 1141 | 31.85 |
| 1145 | 30.37 |
| 1149 | 31.24 |
| 1159 | 31.34 |
Plate 11: SDR 1; SDR 641; 27°C; 20221026
Time start: 12:29
Time R start: 12:49
Time end: 13:04
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | B1 | B2 | B3 | A1 | A2 | A3 |
| B | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| C | B1 | B2 | B3 | A1 | A2 | A3 |
| D | B1 | B2 | B3 | A1 | A2 | A3 |
Well A1: 4 larvae
Well A2: 4 larvae
Well C3: 3 larvae
Well D1: 4 larvae
At the end of this run, some larvae had escaped from the wells. We will fix this by sealing the coverslips more securely on the next run. The number of larvae escaped were:
1 out of A1
1 out of A3
1 out of C3
1 out of D1
1 out of D4
3 out of D5
3 out of D6
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1229 | 28.34 |
| 1251 | 28.26 |
| 1258 | 27.82 |
Plate 12: SDR 2; SDR 710; 30°C; 20221026
Time start: 12:29
Time R start: 12:49
Time end: 13:04
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | B1 | B2 | B3 | A1 | A2 | A3 |
| B | B1 | B2 | B3 | A1 | A2 | A3 |
| C | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| D | B1 | B2 | B3 | A1 | A2 | A3 |
Well B3: 4 larvae
Well B6: 4 larvae
At the end of this run, some larvae had escaped from the wells. We will fix this by sealing the coverslips more securely on the next run. The number of larvae escaped were:
3 out of A3
3 out of A4
2 out of A5
1 out of B6
2 out of B2
1 out of D4
| Temperature measurements | |
|---|---|
| Time | Temp |
| 1229 | 31.41 |
| 1251 | 29.99 |
| 1258 | 30.12 |
- 3. Sampling for metabolomics and RNA/DNA: Larvae were sampled today using the same protocol as on 24 October 2022. Larvae were counted from tanks in random order and placed in an incubator at the treatment temperature (either 27°C or 30°C) for 10-15 minutes prior to sampling. Larvae in falcon tubes were kept in the treatment tanks (i.e., a water bath) until counted. After counting, remaining larvae were poured back into the respective treatment tank.
The tubes sampled today were:
R37-R72 (n=3 tubes per tank, n=50 larvae per tube)
M37-M72 (n=3 tubes per tank, n=50 larvae per tube)
| Tube ID | Date | Time | Tank | Number larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| M37 | 20221026 | 1717 | A1 | 50 | Flash frozen | -40C |
| M38 | 20221026 | 1717 | A1 | 50 | Flash frozen | -40C |
| M39 | 20221026 | 1717 | A1 | 50 | Flash frozen | -40C |
| M40 | 20221026 | 1406 | A2 | 50 | Flash frozen | -40C |
| M41 | 20221026 | 1406 | A2 | 50 | Flash frozen | -40C |
| M42 | 20221026 | 1406 | A2 | 50 | Flash frozen | -40C |
| M43 | 20221026 | 1445 | A3 | 50 | Flash frozen | -40C |
| M44 | 20221026 | 1445 | A3 | 50 | Flash frozen | -40C |
| M45 | 20221026 | 1445 | A3 | 50 | Flash frozen | -40C |
| M46 | 20221026 | 1630 | A4 | 50 | Flash frozen | -40C |
| M47 | 20221026 | 1630 | A4 | 50 | Flash frozen | -40C |
| M48 | 20221026 | 1630 | A4 | 50 | Flash frozen | -40C |
| M49 | 20221026 | 1542 | A5 | 50 | Flash frozen | -40C |
| M50 | 20221026 | 1542 | A5 | 50 | Flash frozen | -40C |
| M51 | 20221026 | 1542 | A5 | 50 | Flash frozen | -40C |
| M52 | 20221026 | 1655 | A6 | 50 | Flash frozen | -40C |
| M53 | 20221026 | 1655 | A6 | 50 | Flash frozen | -40C |
| M54 | 20221026 | 1655 | A6 | 50 | Flash frozen | -40C |
| M55 | 20221026 | 1705 | B1 | 50 | Flash frozen | -40C |
| M56 | 20221026 | 1705 | B1 | 50 | Flash frozen | -40C |
| M57 | 20221026 | 1705 | B1 | 50 | Flash frozen | -40C |
| M58 | 20221026 | 1350 | B2 | 50 | Flash frozen | -40C |
| M59 | 20221026 | 1350 | B2 | 50 | Flash frozen | -40C |
| M60 | 20221026 | 1350 | B2 | 50 | Flash frozen | -40C |
| M61 | 20221026 | 1530 | B3 | 50 | Flash frozen | -40C |
| M62 | 20221026 | 1530 | B3 | 50 | Flash frozen | -40C |
| M63 | 20221026 | 1530 | B3 | 50 | Flash frozen | -40C |
| M64 | 20221026 | 1435 | B4 | 50 | Flash frozen | -40C |
| M65 | 20221026 | 1435 | B4 | 50 | Flash frozen | -40C |
| M66 | 20221026 | 1435 | B4 | 50 | Flash frozen | -40C |
| M67 | 20221026 | 1606 | B5 | 50 | Flash frozen | -40C |
| M68 | 20221026 | 1606 | B5 | 50 | Flash frozen | -40C |
| M69 | 20221026 | 1606 | B5 | 50 | Flash frozen | -40C |
| M70 | 20221026 | 1640 | B6 | 50 | Flash frozen | -40C |
| M71 | 20221026 | 1640 | B6 | 50 | Flash frozen | -40C |
| M72 | 20221026 | 1640 | B6 | 50 | Flash frozen | -40C |
| Tube ID | Date | Time | Tank | Number larvae | Sample method | Storage |
|---|---|---|---|---|---|---|
| R37 | 20221026 | 1717 | A1 | 50 | RNA DNA Shield 600uL | -40C |
| R38 | 20221026 | 1717 | A1 | 50 | RNA DNA Shield 600uL | -40C |
| R39 | 20221026 | 1717 | A1 | 50 | RNA DNA Shield 600uL | -40C |
| R40 | 20221026 | 1406 | A2 | 50 | RNA DNA Shield 600uL | -40C |
| R41 | 20221026 | 1406 | A2 | 50 | RNA DNA Shield 600uL | -40C |
| R42 | 20221026 | 1406 | A2 | 50 | RNA DNA Shield 600uL | -40C |
| R43 | 20221026 | 1445 | A3 | 50 | RNA DNA Shield 600uL | -40C |
| R44 | 20221026 | 1445 | A3 | 50 | RNA DNA Shield 600uL | -40C |
| R45 | 20221026 | 1445 | A3 | 50 | RNA DNA Shield 600uL | -40C |
| R46 | 20221026 | 1630 | A4 | 50 | RNA DNA Shield 600uL | -40C |
| R47 | 20221026 | 1630 | A4 | 50 | RNA DNA Shield 600uL | -40C |
| R48 | 20221026 | 1630 | A4 | 50 | RNA DNA Shield 600uL | -40C |
| R49 | 20221026 | 1542 | A5 | 50 | RNA DNA Shield 600uL | -40C |
| R50 | 20221026 | 1542 | A5 | 50 | RNA DNA Shield 600uL | -40C |
| R51 | 20221026 | 1542 | A5 | 50 | RNA DNA Shield 600uL | -40C |
| R52 | 20221026 | 1655 | A6 | 50 | RNA DNA Shield 600uL | -40C |
| R53 | 20221026 | 1655 | A6 | 50 | RNA DNA Shield 600uL | -40C |
| R54 | 20221026 | 1655 | A6 | 50 | RNA DNA Shield 600uL | -40C |
| R55 | 20221026 | 1705 | B1 | 50 | RNA DNA Shield 600uL | -40C |
| R56 | 20221026 | 1705 | B1 | 50 | RNA DNA Shield 600uL | -40C |
| R57 | 20221026 | 1705 | B1 | 50 | RNA DNA Shield 600uL | -40C |
| R58 | 20221026 | 1350 | B2 | 50 | RNA DNA Shield 600uL | -40C |
| R59 | 20221026 | 1350 | B2 | 50 | RNA DNA Shield 600uL | -40C |
| R60 | 20221026 | 1350 | B2 | 50 | RNA DNA Shield 600uL | -40C |
| R61 | 20221026 | 1530 | B3 | 50 | RNA DNA Shield 600uL | -40C |
| R62 | 20221026 | 1530 | B3 | 50 | RNA DNA Shield 600uL | -40C |
| R63 | 20221026 | 1530 | B3 | 50 | RNA DNA Shield 600uL | -40C |
| R64 | 20221026 | 1435 | B4 | 50 | RNA DNA Shield 600uL | -40C |
| R65 | 20221026 | 1435 | B4 | 50 | RNA DNA Shield 600uL | -40C |
| R66 | 20221026 | 1435 | B4 | 50 | RNA DNA Shield 600uL | -40C |
| R67 | 20221026 | 1606 | B5 | 50 | RNA DNA Shield 600uL | -40C |
| R68 | 20221026 | 1606 | B5 | 50 | RNA DNA Shield 600uL | -40C |
| R69 | 20221026 | 1606 | B5 | 50 | RNA DNA Shield 600uL | -40C |
| R70 | 20221026 | 1640 | B6 | 50 | RNA DNA Shield 600uL | -40C |
| R71 | 20221026 | 1640 | B6 | 50 | RNA DNA Shield 600uL | -40C |
| R72 | 20221026 | 1640 | B6 | 50 | RNA DNA Shield 600uL | -40C |
- 4. Dosing symbionts: We airbrushed 5 parent fragments and isolated symbionts as described on 25 October 2022. Half of the isolate was refrigerated for the evening dosing. The dosing information is as follows:
| Date | Time | Total Volume mL | Squares Counted | Count1 | Count2 | Count3 | Count4 | Count5 | Count6 | Mean Count | Cells per mL | Cells total | Notes | Date | Pool | Symbiont Density (C1) | Target Density (C2) | Volume Symbionts (V1) | Tank volume (V2) | Time Added/Water Off | Time Water On | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20221026 | 1735 | 45 | 1 | 223 | 196 | 204 | 210 | 208.25 | 20825000 | 937125000 | 20221026 | 5 parents | 937125000 | 30000 | 0.5762304922 | 18000 | 1740 | 1840 | ||||
| 20221026 | 2100 | 45 | 1 | 223 | 196 | 204 | 210 | 208.25 | 20825000 | 937125000 | 20221026 | 5 parents | 937125000 | 30000 | 0.5762304922 | 18000 | 2100 | 2200 |
0.57 mL of symbiont isolate was added to each symbiont addition tank at 1740 (water off 1740-1840) and at 2100 (water off 2100-2200).
- 5. Symbiont cell counts: 8 larvae from each tank were collected at 2200. There were only a few larvae with 1-2 cells present. Three larvae had 1 cell each from tank A2 and 2 larvae from tank B3 had one cell. No other larvae had visible symbiont cells. In the future a fluorescent microscope would be ideal for this method.
Larval rearing
Daily measurements in all squaricals were collected from 1800-1830.
Tanks were stirred and maintained throughout the day. Temperatures were on target as expected. Filters were cleaned every 4-6 hours. As the filters get dirtier, they need to be cleaned more often. Recommend that filters are replaced before the next month’s spawning for the field team.
25 October 2022
Huffmyer - Dosing symbionts - day 2 of experiment
We will add symbionts twice daily to experimental tank systems for 3 days (72 hours) to provide symbionts and opportunity for symbiont uptake in larvae. Each day, we will freshly isolate symbionts from parent fragments in the morning and refrigerate the symbionts remaining after dosing to dose in the evening. Symbiont isolation and dosing followed this protocol daily:
- Gather 5-6 randomly selected fragements from different colonies of adult A. pulchra colonies (parental colonies).
- Airbrush the fragments into a single ziploc bag to pool and combine all tissue from all fragments.
- The tissue slurry was evenly divided into two 50 mL falcon tubes. One will be used for dosing in the morning and the other will be kept for dosing in the afternoon.
- The slurry was then cleaned to isolate the symbionts. The falcon tubes were spun in a 50 mL tube centrifuge at 4000 rmp for 4 minutes.
- The supernatant was poured off and 25 mL of 0.02 µm FSW was added. The slurry was then homogenized by shaking the tube for ~30 sec.
- Repeat this process 3 times.
- After the last spin and pouring off the supernatant, 40 mL of 0.02 µm FSW was added and the tube was homogenized by shaking. This is the isolated symbiont pool.
- Today, we will split the extra falcon tube into two falcon tubes (~20 mL symbionts per tube) and kept in 1) an incubator in the dark at 27°C and 2) in the dark at 4°C in the fridge. In the evening we will see which culture has remained the highest quality for dosing.
- Next, count symbiont cells to calculate cell density in the symbiont pool and calculate the amount required for dosing. We are aiming for 3 x 10^4 cells per mL as described in Bay et al. 2011 and Hartmann et al. 2018.
- We used a Google Drive spreadsheet to calculate the volume of pool required to achieve the target density. Today’s calculations are as follows.
| Date | Time | Total Volume mL | Squares Counted | Count1 | Count2 | Count3 | Count4 | Count5 | Count6 | Mean Count | Cells per mL | Cells total | Notes | Date | Pool | Symbiont Density (C1) | Target Density (C2) | Volume Symbionts (V1) | Tank volume (V2) | Time Added/Water Off | Time Water On | Notes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20221025 | 1000 | 40 | 1 | 151 | 159 | 171 | 183 | 166 | 16600000 | 664000000 | 20221025 | 5 parents | 664000000 | 30000 | 0.813253012 | 18000 | 1015 | 1133 | |||||
| 20221025 | 2000 | 40 | 1 | 151 | 159 | 171 | 183 | 166 | 16600000 | 664000000 | 20221025 | 5 parents | 664000000 | 30000 | 0.813253012 | 18000 | 1900 | 2000 |
- Today we added 0.81 mL of symbiont pool to tanks A1-3 and B1-3. We also added symbionts to the extra settlement bins. Symbionts were added with a pipette and stirred in the tanks.
- We added symbionts at 1015 and turned water off at this time. Water was turned back on at 1133 to keep the concentration higher before returning flow through water supply.
- In the afternoon, symbionts in the fridge were better suited for infections. The symbionts in the incubators smelled stronger of tissue and to reduce risk of degradation and adding damaged tissue to the larvae, we selected the refrigerated pool to dose in the afternoon/evening.
- The same volume of symbionts was added to tanks at 1900 with water turned off until 2000.
Huffmyer - Cell counts from 24 October 2022 larvae
Ariana counted cell density in larvae collected yesterday for the initial time point between 1100 and 1300. The larvae all survived in the incubators at 27°C in the dark. The protocol for cell counts was as follows:
- Three larvae were added in a drop of water on a glass slide (load on the left, center, and right areas of the slide).
- A glass coverslip was added on top of each larvae and pressed down firmly to “squish” the larvae.
- The slide was loaded onto a compound microscope and visualized at 20 and 40X.
- Scroll through the tissue to look for symbionts.
- Once all cells are counted in the three larvae loaded, clean the slide with DI water followed by an alcohol wipe.
- Look at the slide under the scope to ensure any tissue symbiont cells are not present after cleaning.
- Repeat until all larvae are examined.
Today we obesrved 0 symbiont cells in all larvae (n=8 larvae per tank, N=96 larvae). We visualized larvae and cells on a stage micrometer (1 DIV = 0.01 mm) to understand the size and characteristics of symbiont cells to train ourselves.
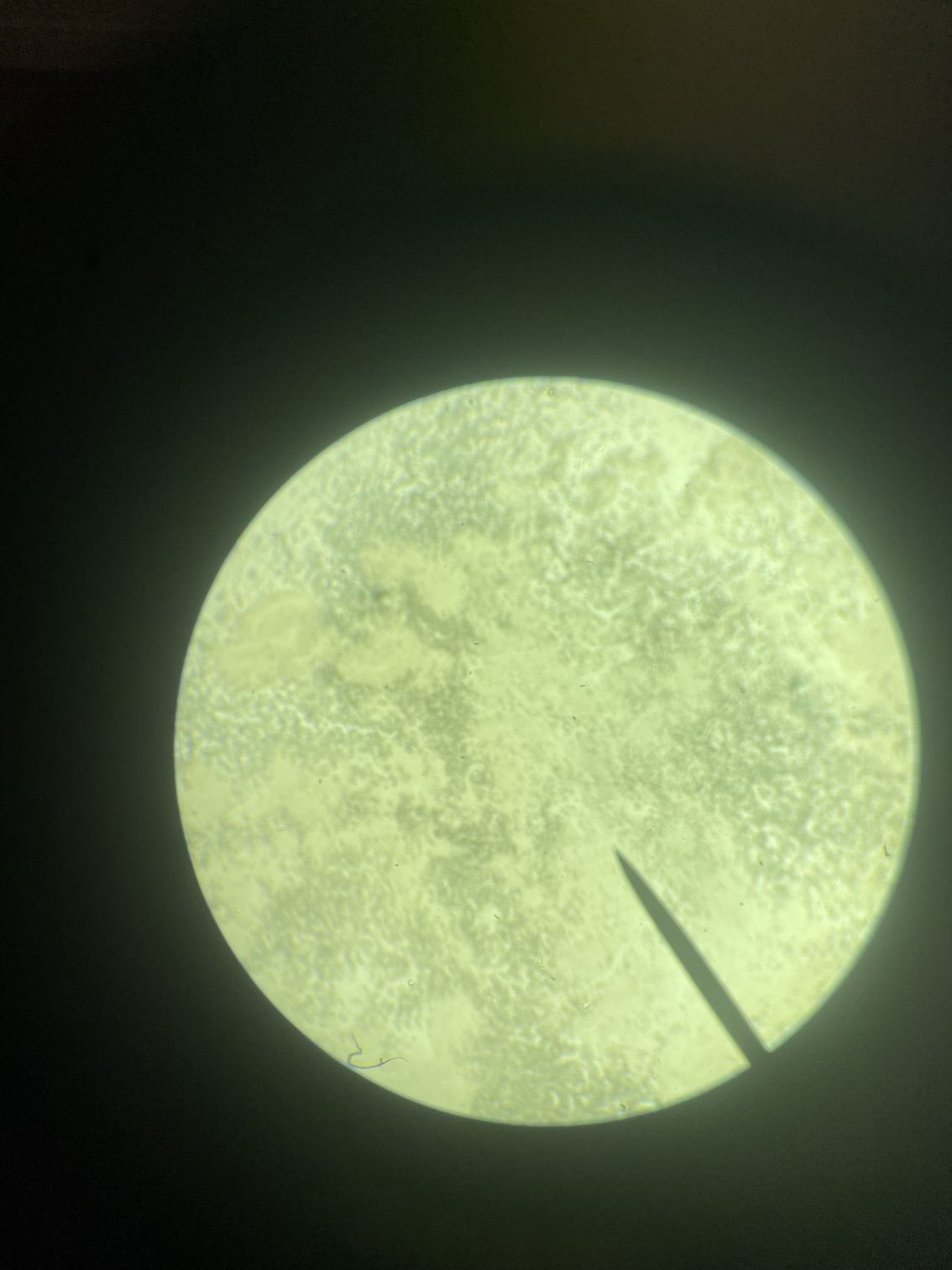
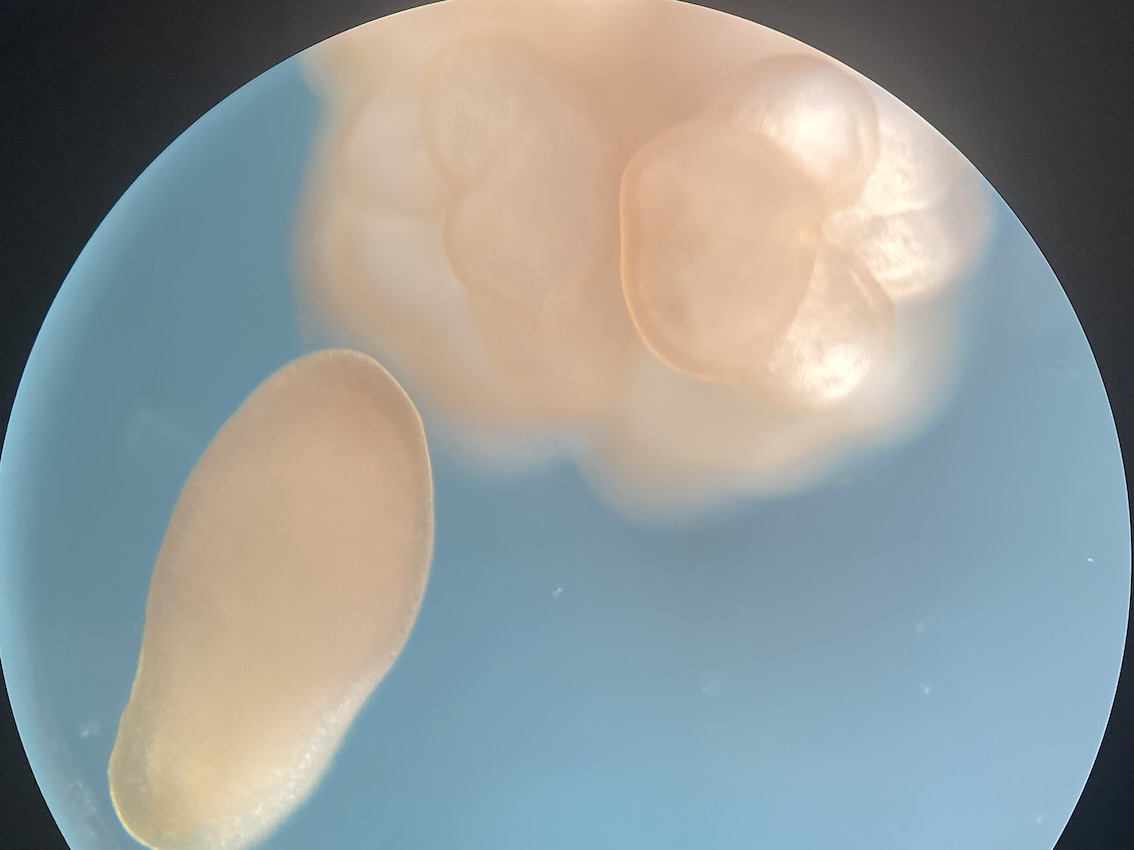
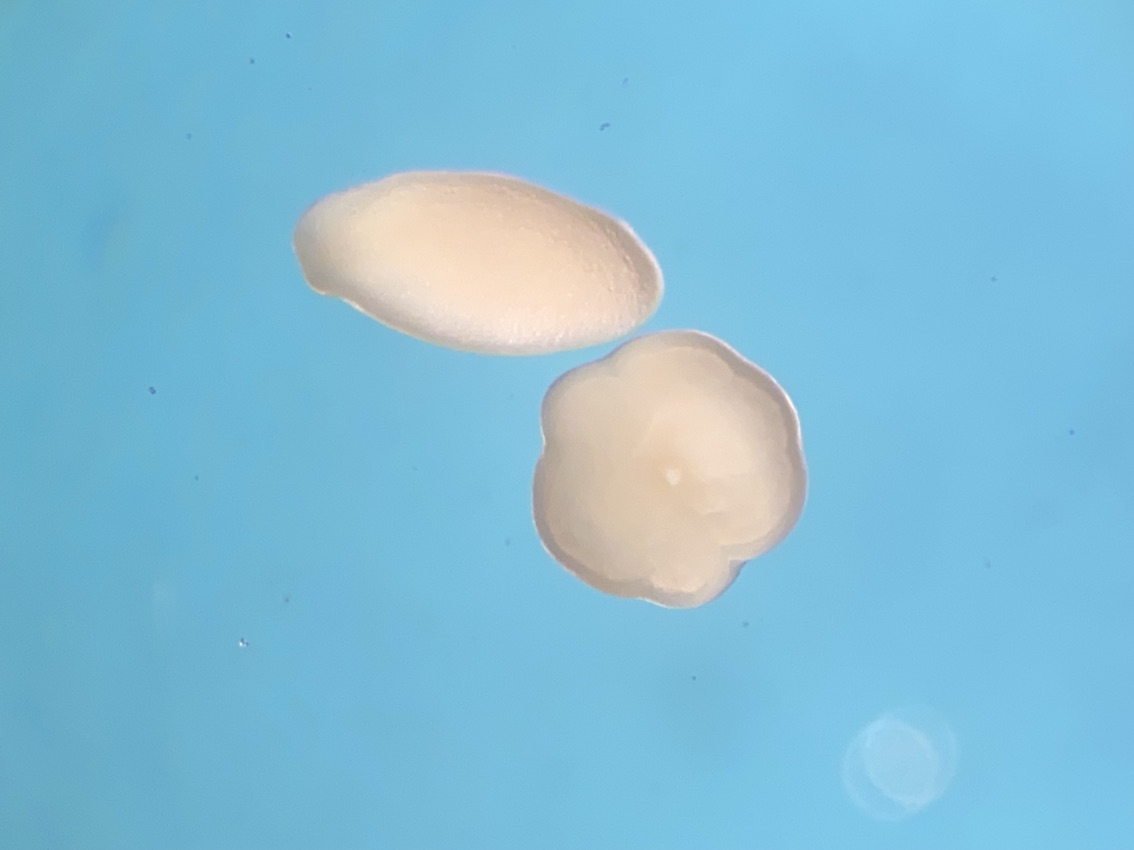
Here is an example of one larave squished under a cover slip on a glass slide.

On the compound microscope, this is what the squished larval tissue looks like. This sample had 0 symbiont cells.
We also loaded some of the isolated symbionts onto the slide to visualize what the cells look like. The cells were 5-10 µm in diameter and had characteristic structure and coloration.
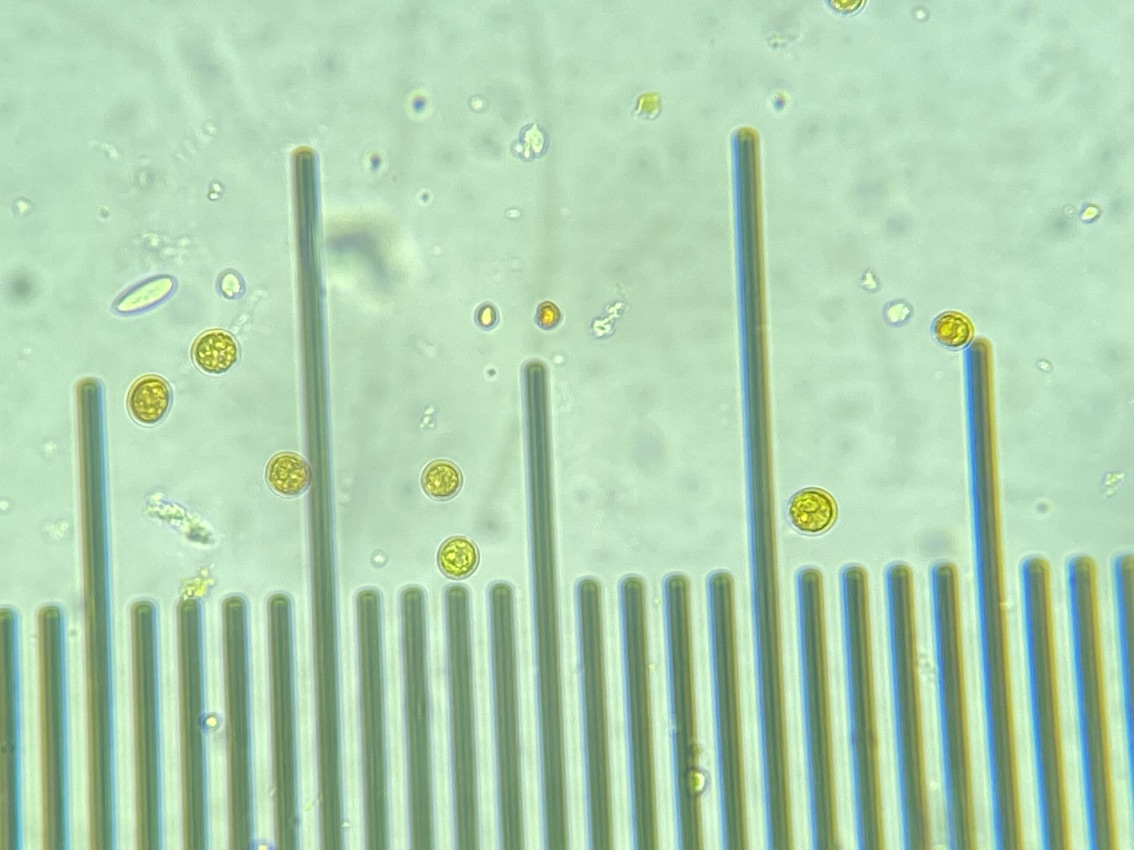
We also loaded a slide with symbiont cells next to a live larvae for size comparison.

We will take n=8 larave for cell counting on Wednesday afternoon (mid point) as well as Friday morning (day of final sampling).
Visualizing larvae under the microscope
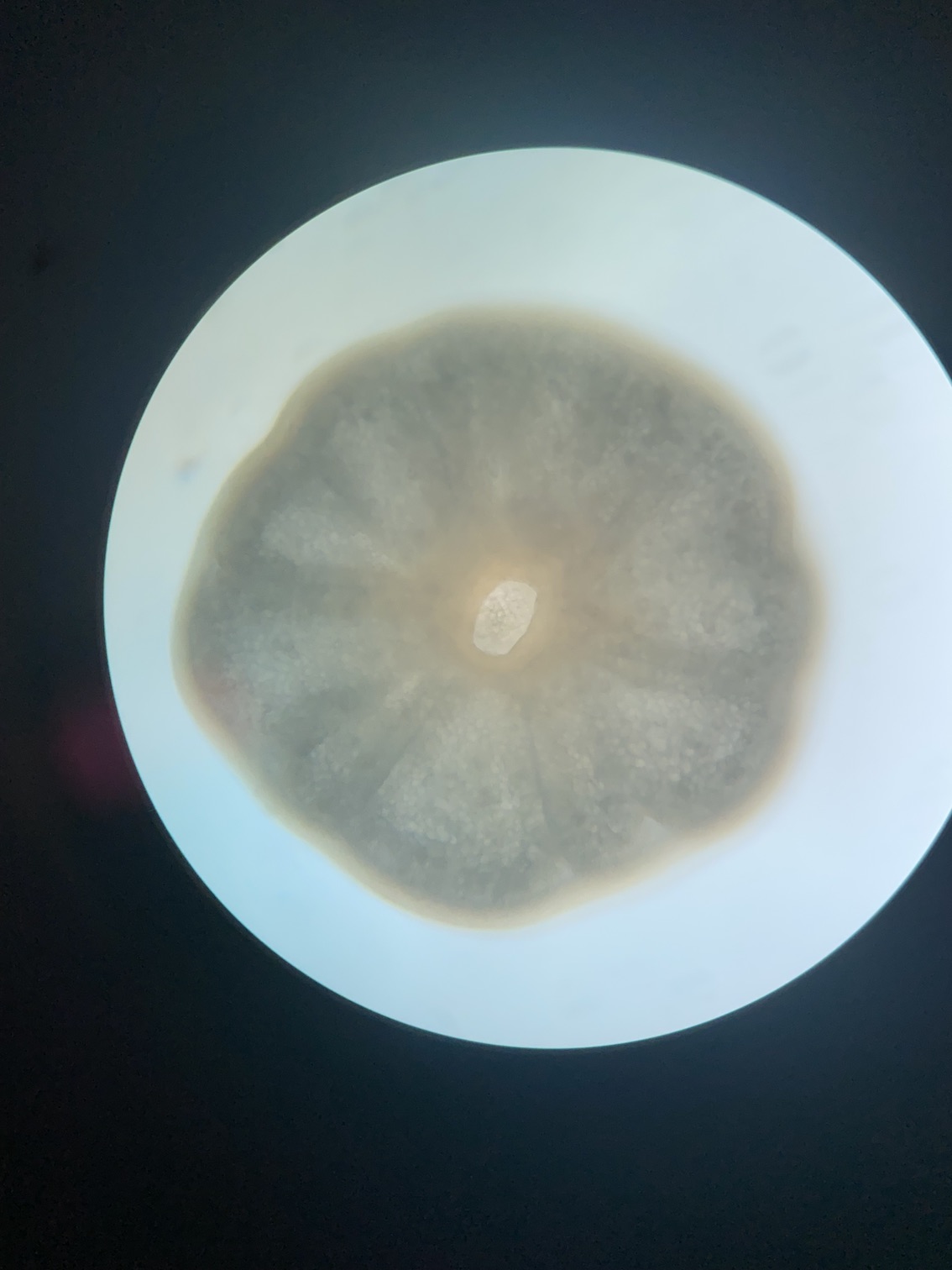
We took some time today to visualize larvae under the microscope. We observed clear swimming and cilia movement behavior and the oral pore was developed in all larvae that we looked at, consistent with our expectations of developmental time points.
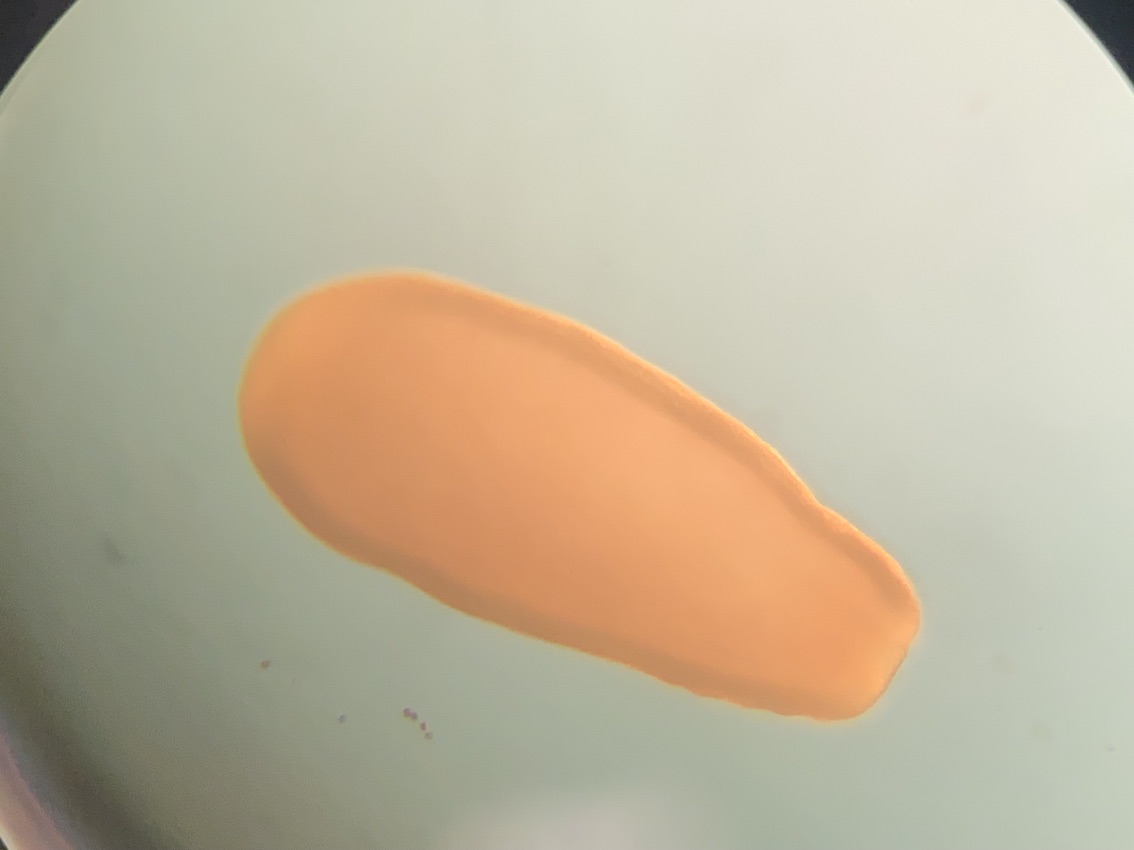
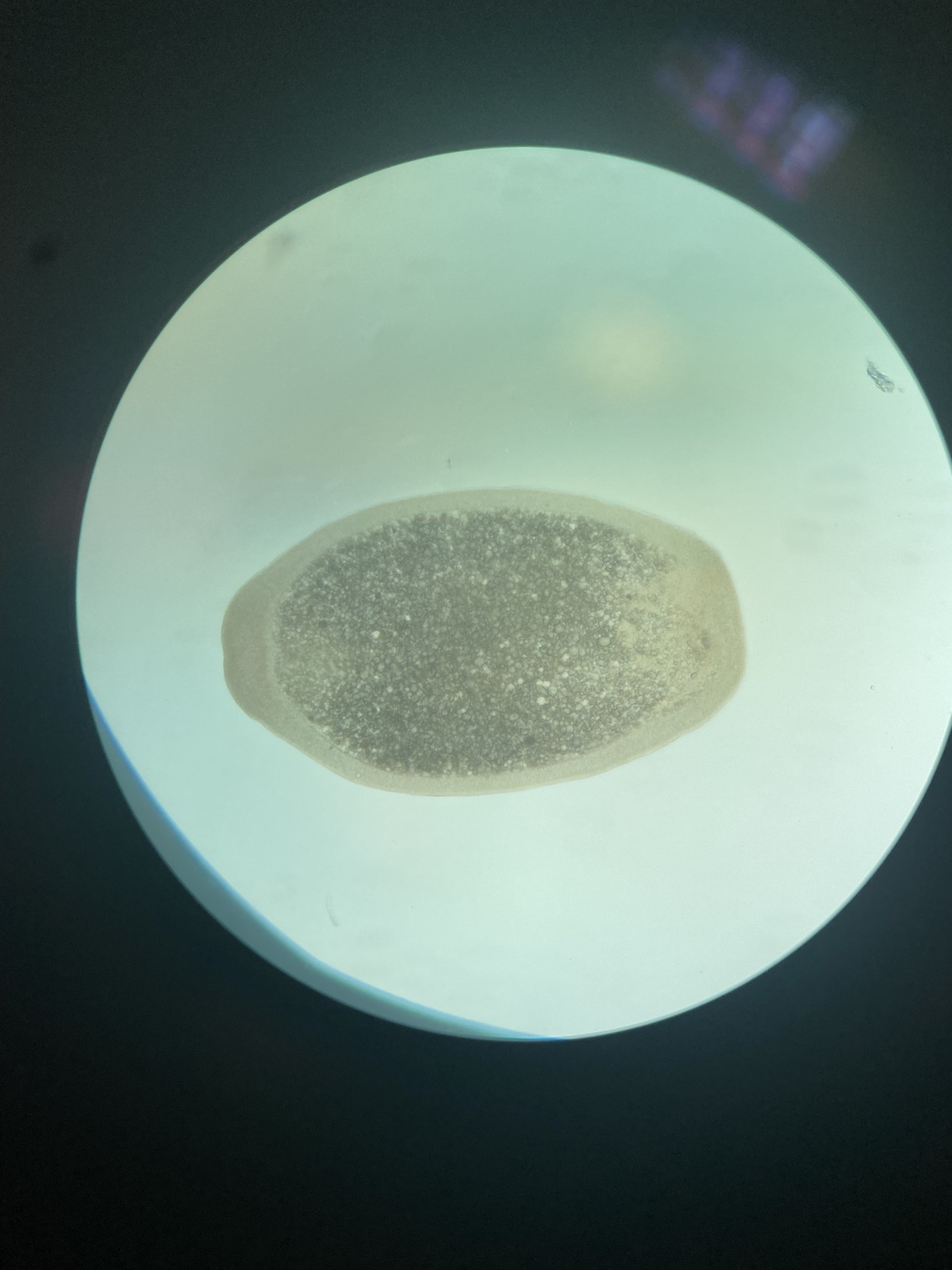
We also visualized larvae next to metamorphosed polyps. We noticed that about half of individuals are at the larval stage and half at the metamorphosed stage in all tanks.
Huffmyer - Larval rearing
We cleaned all coolers and tubing between 1600 and 1730 today. We also cleaned all larval tanks and measured larval densities. We will not be able to use larval density to monitor survival due to high rates of settlement that confound density measurements. Rather, we will use density measurements to make sure we have enough larvae for each sampling time point.
We carefully rinsed all material between symbiont dosing and no dosing tanks to avoid any contamination of symbionts into the no dosing tanks.
Larvae from each tank were poured through a 153 µm seive and then rinsed into a tripour with volume markings. From this pool, 6 1 mL samples were taken and added into a 6-well plate. Larvae were counted in each well to generate larval density measurements in the table below for each tank. After pooling and measuring density, tanks were cleaned, refilled, and larvae were added back to each respective tank.
| Tank | Time | Pool Volume | Rep 1 | Rep 2 | Rep 3 | Rep 4 | Rep 5 | Rep 6 | Rep Volume mL | Larvae per mL | Total Larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 1720 | 400 | 3 | 2 | 6 | 2 | 6 | 5 | 1 | 4 | 1600 |
| A2 | 1730 | 350 | 8 | 9 | 10 | 11 | 9 | 4 | 1 | 8.5 | 2975 |
| A3 | 1735 | 450 | 9 | 13 | 7 | 4 | 6 | 3 | 1 | 7 | 3150 |
| A4 | 1746 | 550 | 8 | 10 | 3 | 7 | 8 | 3 | 1 | 6.5 | 3575 |
| A5 | 1753 | 400 | 8 | 5 | 8 | 10 | 7 | 2 | 1 | 6.6 | 2666 |
| A6 | 1759 | 400 | 5 | 7 | 3 | 2 | 5 | 6 | 1 | 4.6 | 1866 |
| B1 | 1810 | 500 | 2 | 2 | 4 | 7 | 5 | 10 | 1 | 5 | 2500 |
| B2 | 1818 | 620 | 4 | 4 | 2 | 0 | 5 | 4 | 1 | 2.5 | 1550 |
| B3 | 1825 | 400 | 6 | 8 | 7 | 6 | 5 | 5 | 1 | 6.6 | 2666 |
| B4 | 1835 | 520 | 4 | 2 | 7 | 5 | 4 | 6 | 1 | 4.6 | 2426 |
| B5 | 1838 | 650 | 5 | 3 | 4 | 4 | 3 | 5 | 1 | 4.2 | 2704 |
| B6 | 1843 | 300 | 5 | 13 | 4 | 3 | 5 | 3 | 1 | 5.5 | 1650 |
On average, each tank had 2,444 larvae, which is reduced from 4,259 estimated as the starting density. There is still enough for sampling with some mortality or settlement. The lowest density is in tank B2 (1,550) and the highest was A4 (3,575).
Huffmyer - Temperature treatments
Temperature treatments are tracking as expected. Apex was monitored throughout the day and temperature loggers will be downloaded on Friday at the end of the experiment.
Water flow was increased to 20-25 mL per 5 seconds in each tank to help the temperature stabilize and reduce variability in each tank.
Daily measurements were taken by Ariana and Terava in all larval tanks at 1520-1540.
E5 experiment
Daily measurements were taken by Terava and Ariana in the E5 tanks today.
Calibrating salinity probe
We were able to successfully calibrate the salinity probe today. It has been reading high due to a bad calibration solution. Hollie calibrated to a 1000 µS/cm YSI solution successfully with tank measurements reading 35-36 psu as expected. THe average cell cal factor was 0.457 after calibration at 1413. We will need to apply a correction to all values before this date.
24 October 2022
Huffmyer - Initial sampling on day 1 of experiment
Today is the first day of our A. pulchra larval temperature experiment! We are going to conduct sampling today prior to the initiation of temperature exposure. After sampling is complete, we will start temperature treatment. Larvae will be exposed to temperature treatment (elevated ~1.5°C above ambient) for 5 days (through Friday) with sampling on days 3 and 5.
Details of sampling are described below for today in the order listed.
-
1. Obtain larvae from each squarical: We need to have ~400 larvae from each squarical (n=12 squaricals). This will be used for RNA/DNA sampling (n=50 larvae per squarical), metabolomics sampling (n=50 larvae per squarical), respirometry (n=30 larvae per squarical), and cell density measurements (n=8 larvae per squarical). Larvae were obtained by scooping water out of each squarical through a seive (153 µm mesh bottom bin) and then pipetted/rinsed into a 50 mL falcon tube labeled for each squarical. These falcon tubes were then kept in the treatment tanks during sampling and brought to the lab in turn for each to be sampled in random order.
-
2. Respirometry: Respirometry measurements followed Ariana’s larval respirometry protocol.
- Both the 710 (SDR 2) and the 641 (SDR 1) plates were hydrated and then loaded with 100% air saturated 0.02 µm filtered seawater bubbled for 3-5 min (FSW). We allowed the plates to come to treatment temperature (27°C for the 641 plate and 30°C for the 710 plate) in benchtop MyTemp incubators. The plates were then calibrated (one point calibration in SDR software) at the treatment temperature using 100% air saturated FSW as the calibration standard. One point calibration at 0815.
- A digital thermometer was used to continuously monitor temperature throughout all respiration runs.
- Hollie and Ariana counted larvae into groups of 5 larvae per well. The measurements followed the plate maps listed below.
- Temperature, plate maps, and light measurements were recorded in a Google Drive spreadsheet. This data will be moved to GitHub in the coming weeks.
- AquaIllumination Hydra 16 HD lights were set to 30% intensity on all channels to generate light at ~500 PAR at the level of the respirometry plate. At the beginning of each day, light was measured in the center and in each corner of the plate and recorded in the Google Drive spreadsheet linked above.
- In between runs, falcon tubes of larvae were kept in the treatment tanks to avoid effects of changing temperature indoors. All larvae were kept in the lab for the same amount of time during the counting process before each run.
- After counting into groups of 5 larvae, larvae were loaded into the SDR wells at the specified positions. Wells were filled with 0.02 µm FSW and sealed with glass coverslips as described in the respirometry protocol. If larvae escaped during the sealing process the number of larvae remaining in each well was noted in the Google Drive spreadsheet.
- After loading, SDR 1 (641) and SDR 2 (710) were added into incubators at 27°C or 30°C, respectively. The data collection was then started in the software. Measurements were collected every 15 sec (salinity = 36, pressure = 965, batch = PSt 1624, units = % air saturation) for 20-25 minutes under the light (photosynthesis, if photosynthesis is active), and in the dark (light-enhanced respiration).
- The light measurements are run first to standardize the light environment prior to respiration measurements. Light measurements were made to monitor photosynthesis, because a treatment in this experiment is symbiont infection (see information from 23 October 2022 on experimental design).
Plate maps for respirometry today
The plate maps are included below along with information on plate run times and temperature treatments. In all plate maps, 5 larvae were loaded into each well. The information inside each well indicates the squarical tank that the larvae came from. “A” tanks are high temperature and “B” tanks are ambient. Tanks 1-3 will be dosed with symbionts whereas tanks 4-6 will not be dosed with symbionts.
Plate 1: SDR 1 - 641; 27°C; 20221024
Time start: 8:53
Time respiration start: 9:18
Time respiration ends: 9:33
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| B | A1 | A2 | A3 | B1 | B2 | B3 |
| C | A1 | A2 | A3 | B1 | B2 | B3 |
| D | A1 | A2 | A3 | B1 | B2 | B3 |
Well D1: 6 larvae
Plate 2: SDR 2 - 710; 30°C; 20221024
Time start: 8:53
Time respiration start: 9:18
Time respiration ends: 9:33
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A1 | A2 | A3 | B1 | B2 | B3 |
| B | A1 | A2 | A3 | B1 | B2 | B3 |
| C | A1 | A2 | A3 | B1 | B2 | B3 |
| D | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
Well B1: 4 larvae
Well B2: 4 larvae
Plate 3: SDR 1 - 641; 27°C; 20221024
Time start: 10:25
Time respiration start: 10:50
Time respiration ends: 11:05
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A4 | A5 | A6 | B4 | B5 | B6 |
| B | A4 | A5 | A6 | B4 | B5 | B6 |
| C | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| D | A4 | A5 | A6 | B4 | B5 | B6 |
Well A6: 4 larvae
Plate 4: SDR 2 - 710; 30°C; 20221024
Time start: 10:25
Time respiration start: 10:50
Time respiration ends: 11:05
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | A4 | A5 | A6 | B4 | B5 | B6 |
| B | BLANK | BLANK | BLANK | BLANK | BLANK | BLANK |
| C | A4 | A5 | A6 | B4 | B5 | B6 |
| D | A4 | A5 | A6 | B4 | B5 | B6 |
Well C3: 3 larvae
Well D3: 4 larvae
During all measurements blanks appeared to have minimal slope as expected. We also noticed that during the light phase there were no positive slopes indicating no photosynthesis. This is expected with aposymbiotic larvae. Respiration slopes during the dark phase were negative as expected with oxygen consumption.
Each day of sampling, we will alternate the order of which tanks are run in the first set of plates to avoid any timing effects.
-
3. Sampling for metabolomics and RNA/DNA:
- After respiration measurements were completed, we sampled larvae for metabolomics (untargetted) and RNA/DNA (gene expression). Sampling followed this protocol:
- All falcon tubes with larvae were kept in the treatment tanks until brought in for counting. We randomly selected one falcon tube (i.e., larvae from one tank) of the 12 tubes (12 tanks) to bring into the lab at a time. Hollie then counted n=6 groups of 50 larvae and placed in a 6-well plate.
- To standardize the environment prior to sampling after counting, larvae were then kept in an incubator at ~200 PAR (10% all channels) for 10-15 minutes at the treatment temperature (either 27°C for B tanks or 30°C for A tanks) prior to sampling.
- After incubation, the larvae were then put into 2 mL screw top cryo vials by Ariana with a pipette and sampled.
- Larvae were sampled either for metabolomics (“M” tubes) or RNA/DNA (“R” tubes). Tubes were pre-labeled with the letter prefix followed by a number.
- For each larval tank, n=3 samples of 50 larvae were taken for metabolomics and n=3 samples were taken for RNA/DNA.
- Sample ID’s were recorded in Google Drive. Date and time of sampling were recorded in the Google Drive sheet.
- To sample, larvae were taken out of the 6- well plate with a glass pipette and added into labeled tubes. Each well of n=50 larvae was added into one tube. Tubes were then spun in a bench top centrifuge for 1.5 min at 13,000g.
- Three tubes were spun at a time (e.g., all M tubes then all R tubes for each tank).
- After spinning, as much seawater was removed as possible with a glass pipette.
- M tubes were then directly flash frozen in liquid nitrogen and then stored in a box labeled with A. Huffmyer and project information and kept at -40°C in the molecular lab freezer.
- R tubes were sampled by adding 600 µL of RNA DNA Shield directly to the tube. After all tubes were sampled, the R tubes were placed in the -40°C freezer in a box at the end of the day.
The tubes sampled today were:
R1-R36 (n=3 tubes per tank, n=50 larvae per tube)
M1-M36 (n=3 tubes per tank, n=50 larvae per tube)
During sampling we noticed that about half of the larvae were metamorphosing into the disc/stage 2 phase. Samples contain a mix of swimming and metamorphosed larvae due to limitations in the number of larvae.
Sampling occured between 1229 and 1559.
After sampling all unused larvae were poured back into larval tanks.
Huffmyer - Starting temperature treatment
The treatment breakdown for all tanks is below.
| Tank | Symbiont Treatment | Temperature Treatment |
|---|---|---|
| A1 | Added symbionts | High (+2-2.5°C) |
| A2 | Added symbionts | High (+2-2.5°C) |
| A3 | Added symbionts | High (+2-2.5°C) |
| A4 | No symbionts | High (+2-2.5°C) |
| A5 | No symbionts | High (+2-2.5°C) |
| A6 | No symbionts | High (+2-2.5°C) |
| B1 | Added symbionts | Ambient |
| B2 | Added symbionts | Ambient |
| B3 | Added symbionts | Ambient |
| B4 | No symbionts | Ambient |
| B5 | No symbionts | Ambient |
| B6 | No symbionts | Ambient |
Temperature treatment profiles
Temperature treatment was started at 1723 with target temperature reached by 1900. The treatment was programmed to approximate a 2-2.5°C increase in temperature above ambient conditions. Ambient conditions currently range between 26.75 and 28.0°C daily fluctuation. The target temperature treatment will raise this 2°C to reach a daily fluctuation of 28.75-30.0°C.
In the Apex system, virtual outlet scripts are programmed to turn on 2 800W finnex titanium heaters in the cooler/sump to reach a target temperature for chunks of time during the day to generate a daily cycle in temperature. The full script can be found below.
High temperature treatment profile
28.6C - 30.0C (with 1.5C daily fluctuation)
Stage 1 = 28.6
Stage 2 = 28.8
Stage 3 = 29.0
Stage 4 = 29.2
Stage 5 = 29.4
Stage 6 = 29.6
Stage 7 = 29.8
Stage 8 = 30.0
For high treatment probe with 8 stages
Set OFF
If Outlet High_Stage1 = ON Then ON
If Outlet High_Stage2 = ON Then ON
If Outlet High_Stage3 = ON Then ON
If Outlet High_Stage4 = ON Then ON
If Outlet High_Stage5 = ON Then ON
If Outlet High_Stage6 = ON Then ON
If Outlet High_Stage7 = ON Then ON
If Outlet High_Stage8 = ON Then ON
High_Stage1
Set OFF
If High_1 < 28.6 Then ON
If High_1 > 28.6 Then OFF
If Time 01:00 to 04:30 Then OFF
If Time 06:45 to 00:59 Then OFF
High_Stage2
Set OFF
If High_1 < 28.8 Then ON
If High_1 > 28.8 Then OFF
If Time 01:00 to 03:00 Then OFF
If Time 04:30 to 06:45 Then OFF
If Time 08:15 to 00:59 Then OFF
High_Stage3
Set OFF
If High_1 < 29.0 Then ON
If High_1 > 29.0 Then OFF
If Time 01:00 to 02:00 Then OFF
If Time 03:00 to 08:15 Then OFF
If Time 09:45 to 00:59 Then OFF
High_Stage4
Set OFF
If High_1 < 29.2 Then ON
If High_1 > 29.2 Then OFF
If Time 02:00 to 09:45 Then OFF
If Time 11:15 to 00:30 Then OFF
High_Stage5
Set OFF
If High_1 < 29.4 Then ON
If High_1 > 29.4 Then OFF
If Time 01:00 to 11:15 Then OFF
If Time 12:15 to 23:00 Then OFF
If Time 00:30 to 00:59 Then OFF
High_Stage6
Set OFF
If High_1 < 29.6 Then ON
If High_1 > 29.6 Then OFF
If Time 01:00 to 12:15 Then OFF
If Time 13:30 to 19:45 Then OFF
If Time 23:00 to 00:59 Then OFF
High_Stage7
Set OFF
If High_1 < 29.8 Then ON
If High_1 > 29.8 Then OFF
If Time 01:00 to 13:30 Then OFF
If Time 15:00 to 17:15 Then OFF
If Time 19:45 to 00:59 Then OFF
High_Stage8
Set OFF
If High_1 < 30.0 Then ON
If High_1 > 30.0 Then OFF
If Time 01:00 to 15:00 Then OFF
If Time 17:15 to 00:59 Then OFF
The 800W heaters heat the cooler to the temperature necessary to achieve the desired temperature profile in the larval tanks. Three apex temperature probes were placed in A tanks and B tanks with the temperature profile registering feedback and monitoring one of the larval A tanks. The location of the probes was randomized each day so that the temperature profile is even throughout all the tanks.
The Neptune Apex units used to control this system with high temperature manipulation in A tanks included:
1 Apex base unit (WiFi model)
1 Apex EB832 energy bar
5 PM1 modules
6 Apex temperature probes
Apex aquabus cables between each unit/module
The manipulated header tank then delivered heated water to each larval tank through the manifold delivery system. For more information on the tank systems, refer to the SMILE systems parts list and protocol.
There were no problems with temperature treatment today. The temperature reached the target temperature within ~2 hours and stayed at the correct temperature overnight.
Temperature is monitored in n=3 pendant loggers in A tanks and n=3 pendant loggers in B tanks (A1, A3, A5 and B2, B4, and B6) with logging every 10 min. These loggers were previously calibrated on 25 September 2022. Temperature is monitored live in the Apex Neptune system regularly.
Huffmyer - Sampling for cell counts
Larvae were sampled at 17:50 today to measure symbiont cell densities as an initial time point to see whether larvae had already acquired symbionts in our rearing systems. 8 larvae from each larval tank were collected directly from the tank and loaded into a 96-well plate (n=8 wells per tank). These were kept in an incubator in 0.02 µm FSW at 27°C overnight and will be counted tomorrow morning.
Larval rearing
We collected daily measurements (temperature, pH, salinity, flow) on all larval tanks at 17:07 by Ariana and Danielle. Apex probes were calibrated at 17:00.
Throughout the day, larvae were mixed and tanks were checked every 2-3 hours. Filters were cleaned every 5-7 hours during the day and night. Tanks were checked every 3-4 hours at night.
We noticed continued settlement behavior in larvae. This will impact how many larvae we have at the end of the experiment and will confound larval density measurements for survival.
23 October 2022
Embryo rearing
Today we maintained larval cultures and Danielle cleaned all her squaricals. All of Ariana’s squaricals were also cleaned during pooling/allocating (see below). Filter lines were cleaned three times throughout the day.
We noticed that starting yesterday larvae from 10/18 were starting to settle and stick to the squarical walls at the water line. We are using a pipette to gently brush them off regularly to prevent settling. This is interesting for future experiments to know that this species can settle rapidly.
Symbiont innoculation test
We checked larvae under the microscope by 1) viewing larvae on a scope and 2) squishing larvae under a cover slip to look for cells. No cells were seen in larvae that were exposed to symbionts yesterday.
Experimental design
Due to settlement and no uptake of symbionts, we re-assessed our experimental plan and will proceed with the following design:
All squaricals will be from one cohort. Within eacn large blue tank bath we will have n=3 cultures with symbiont dosing and n=3 cultures without dosing (aposymbiotic vs symbiotic). There will be one blue tank at high temperature (+2C) and one at ambient. We will expose larvae to heat starting tomorrow and sample every other day (Mon, Wed, Fri) in the mornings for 1) metabolomics; 2) gene expression; 3) cell density; and 4) photosynthesis and respiration. At the end of the 5 days, we will proceed with stable isotope labelling incubations for metabolomics if cells are present in the larvae.
This design ensures that even if we do not have larvae take up symbionts, we will investigate a metabolic time series. We will then take the cohort that wasn’t used in the squaricals and place them in settlement bins with settlement plugs.
We will conduct daily symbiont innoculations as described yesterday in the symbiotic tanks (n=6). This design will allow us to test how elevated temperatures impact symbiotic initiation and metabolism.
Today, to move foward with this plan, Ariana pooled and allocated larvae from the 10/19 cohort into 12 squaricals as described yesterday. The 10/18 cohort was added to n=6 settlement bins with 50 aragonite plugs. Settlement bins were mesh bottom bins with the plugs oriented upside down.
Larvae were added at 1730-1745. For 10/18 larvae, there was an average of 27.6 larvae per mL, which resulted in ~4600 larvae in each of the 6 settlement bins. Three bins were kept in tank A and three in tank B partially submerged. Larvae from 10/19 had an average density of 32.3 larvae per mL, which resulted in ~2,691 larvae in each squarical. This number would allow for ~50% mortality and still have enough for sampling.
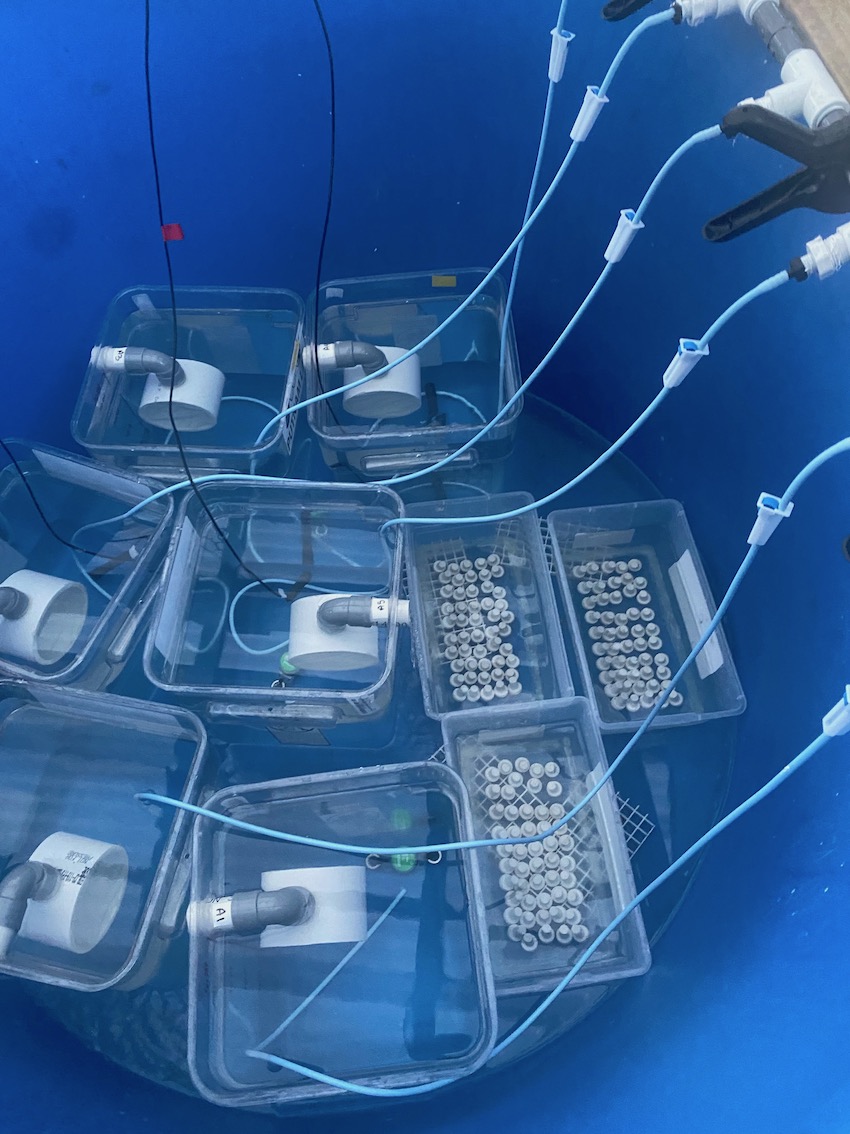
Here is the expected sampling size for this study:
- Metabolomics: n=50 larvae per sample, n=3 samples per squarical; n=3 timepoints; N=450 per squarical
- DNA/RNA: n=50 larvae per sample, n=3 samples per squarical; n=3 timepoints; N=450 per squarical
- Cell density: n=8 larvae per squarical; n=3 timepoints; N=24 per squarical
- Labelling and stable isotopes: n=50 larvae per sample; n=4 samples per squarical; n=1 time point (end); N=200 larvae
- Labelled lipids/MALDI: n=15 larvae per sample; n=3 per squarical; n=1 time point (end)
Daily measurements
Danielle and Hollie took daily measurements today of the E5 tanks and the squaricals between 1500 and 1700.
Spawning
Tonight was a large spawn! It is interesting that the coral’s spawning schedule is not what we expected. It seems the peak is later as it is the 14 night after the full moon. 10-12 of Danielle’s colonies spawned and a large amount of bundles was on the surface of the tank for wildtype. Danielle generated cultures for her project.
The Acropora hyacinthus colonies will be taken back tomorrow, they are unhappy in the tanks.
Preparing for sampling
Ariana labelled well plates for counting and tube boxes for sampling. Tomorrow Ariana will sample for the initial/first time point in the morning.
22 October 2022
Embryo rearing
Buckets and coolers were cleaned today with water changes by pouring water through a mesh container and rinsing back into the clean tank.
At 1600 we looked at larvae under the compound microscope from 10/17. We could clearly see an oral pore.
Sampling - pool and distribute
Today Ariana pooled and distributed all larvae for the symbiont integration project. Squaricals will have n=6 from 10/18 and n=6 from 10/19 for a total of 12 squaricals. The process was as follows.
- Pour water from each squarical through a mesh bottom container in a water bath using a tripour. Take out the mesh and rinse into a Nalgene bottle with volume marks. This is the pool.
- This was done for 10/18 cohort (A1-5, B1-5) and 10/19 cohort (A6, B6, C5, C6, D6). This created two pools of larvae.
- All tanks A1-A6 and B1-B6 were cleaned.
- The concentration of larvae was calculated for each pool at 1230.
For cohort 10/18:
Total volume = 1000 mL of pool. Larvae in 0.5 mL samples of this pool = 12, 15, 22, 20, 18, 16 = 30 larvae per mL. In total this equals about 30,000 in the 10/18 pool.
30,000/6 = approx. 5,000 larvae will be in each squarical
166 mL of the pool will be added to each squarical
Added at 1306.
For cohort 10/19:
Total volume = 1000 mL of pool. Larvae in 0.5 mL samples of this pool = 20, 13, 20, 16, 20, 21 = 36 larvae per mL. In total this equals about 36,000 in the 10/19 pool.
36,000/6 = approx. 6,000 larvae will be in each squarical
166 mL of the pool will be added to each squarical
Added at 1312.
The density of larvae will th3refore be approx. 5,000-6,000 per squarical across cohorts.
Calculating sample needs:
Sampling will use the following amounts of larvae at each sampling from each squarical:
- DNA/RNA = 150 larvae (n=3 reps of 50 larvae)
- Metabolomics = 300 larvae (n=3 reps of 100 larvae)
- P and R = 50 larvae
- Cell density counts = 10 larvae
In total, each sampling will take 510 larvae. Sampling timeline will be described in more detail tomorrow.
At the end of the experiment, we will take samples for stable isotope metabolomics.
- Stable isotope metabolomics = 300 larvae (n=3 reps of 100 larvae)
We will re assess the sampling time line tomorrow with these numbers to decide on sampling time points.
Symbiont innoculations
We conducted a trial of symbiont infections following the protocol from 3 October 2022. We airbrushed one fragment and did 2 cycles of rinsing in 0.02 um FSW and centrifuging in a 50 mL falcon tube at 3500 rpm for 2 min. Ariana then counted cells on the hemocytometer and calculated the volume of symbiont pool to add to one 5 gallon bucket to achieve a density of 3 x 10^4. This volume was 2 mL of slurry. Ariana added this to the 10/17 embryo bucket at 1515. We will look at the larvae tomorrow to see if any mortality occurs or if symbiont uptake occurs. We will use these results to inform our innoculations for Ariana’s project.
Daily measurements
Hollie and Danielle completed daily measurements and cleaned the strings for all of the fragments. During spawning we are also taking daily measurements of squaricals and buckets for reference.
Daily measurements are recorded in GitHub here
Loggers
Ariana downloaded A1-3 and B1-3 pendant loggers at 1230-1300. These were relaunched in new tanks at 1315 as follows:
| SSN | Tank | Notes |
|---|---|---|
| 21002982 | A1 | |
| 21002979 | A3 | |
| 21002974 | A5 | Formerly A2 |
| 21002981 | B2 | Formerly B1 |
| 21335980 | B4 | Formerly B2 |
| 21335987 | B6 | Formerly B3 |
Ariana will analyze these in R the next few days and make scripts to analyze separate from E5.
Spawning
There was much less spawning tonight with 2-4 wildtype in a smaller pool. The pool was fertilized from 2200-2330 and then added to 1 squarical (C5) at 2345.
21 October 2022
High frequency time series
We sampled in the field today for the last high frequency sampling time point (post spawning) at Mahana from 1100-1300. On the trip we also visited the coral restoration tables from the Hay lab. These are great tables that we may consider placing fragments or plugs on in the future. Danielle processed the samples back at the lab.
Embryo rearing
We conducted maintenance of the cultures every 2-3 hours throughout the day and night as previously described for earlier days. We also did water changes in all buckets, coolers, and bins.
We also observed swimming behavior in 10/17 and 10/18 larvae today!
In line water filters were rinsed three times today.
Daily measurements and feeding
Daily measurements were taken in E5 tanks, parent tanks, and larval tanks today between 1530-1630. Daily measurements are recorded in GitHub here. We fed the E5 corals today following the E5 A. pulchra metabolism feeding protocol here. Feeding occurred from 1545-1645.
Sampling prep
Ariana prepared materials and equipment in anticipation of sampling and starting the symbiont integration experiment. Supplies were prepared for:
- Pooling and allocating larvae to treatment tanks
- Symbiont infections/dosing
- Sampling gene expression and metabolomics
- Sampling for symbiont density
- Sampling for respirometry (photosynthesis and respiration)
Sampling will be described in detail when it is completed. The goal is to conduct daily sampling throughout the 1) symbiont infection phase and 2) thermal stress phase.
Spawning
Wildtype colonies 71, 63, and 2 others in the large tank that were not identified produce bundles. The A. hyacinthus colonies do not look good and are experiencing mortality - they need to go back in the field soon.
Spawning for tonight was less than the night before. The largest spawn for wildtype pulchra was 10/19 and 10/20. To make space for Danielle’s fertilization, squaricals A5 and Ay were combined as well as B5 and B6. Then C3 was moved to A6 and D5 was moved to B6 to open space in Danielle’s tanks at 2330. Water changes were done for all buckets and coolers and bins at 2230-2300.
Fertilization and adding embryos to squaricals occured at 0000.
20 October 2022
Embryo rearing
Today we did water changes in all squaricals, buckets, and cultures from 1200-1800. Water was poured through a 153 um large seive. The tanks were cleaned and then the embryos were rinsed back into the tank.
Minimal mortality was observed from last night’s spawning - we did not see much dead material or lipid slick on the surface of the water.
We have noticed that mortailty/degradation of unfertilized eggs occurs between 1200-1400 the day after spawning. This is a time to keep an eye on the cultures and clean if needed. If fertilization success is high, there is very low early mortality.
Coolers were cleaned today and water filter lines were cleaned.
Daily measurements
Daily measurements were taken in E5 tanks, parent tanks, and larval tanks today between 1600 and 1700. Daily measurements are recorded in GitHub here.
Spawning
Wildtype parents that set and released bundles were 74, 75, 77, 72, 64, 71, 80, and 63. Bundles were gathered and fertilized in a pool as described for the previous few days of spawning. Falcon tubes from wildtyp were allocated with 1.5-2 mL of embryos at 2300-0000. In order to make free space in squaricals for Danielle’s fertilization, D3 and D6 squaricals from 10/19 were combined and C4 and C5 were also combined from 10/19.
Wildtype embryos were added to D4 at 2350. There was a lot of extra material, so we found other types of containers to use. We added 0.5-1 mL of bundles to small mesh bins and added 5-8 mL to each of 4 large coolers. Water was changed in all bins in the water table at 2300. Total larvae at this point: 26 squaricals, 6 buckets, 13 bins, 6 tupperware, 4 coolers.
Adult tanks were cleaned at 2100-2200.
19 October 2022
Embryo rearing
Embryos from 10/18 spawning were checked and stirred every 2-3 hours today. We also frequently check the fill level of the coolers. Due to high water flow and limitaitons in rate of filtering in the filter line, the coolers will sometimes drain which risks damaging the pump and stopping water flow. We closely monitor the level of the coolers every 2-3 hours. 50 um in line filters were rinsed and cleaned every 4-7 hours. The coolers were also cleaned today.
Overall, embryos from 10/17 and 10/18 spawning are doing well with minimal mortality.
E5 experiment - feeding
We fed the E5 corals today following the E5 A. pulchra metabolism feeding protocol here. Feeding occurred from 16:35-17:35.
Daily measurements
Daily measurements were taken in E5 tanks, parent tanks, and larval tanks today between 1430 and 1500. Daily measurements are recorded in GitHub here.
In Tank 6, C31 is showing high partial mortality (~90%) with algal overgrowth.
Maintaining water systems
Water lines were flushed and cleaned today and the filters were washed regularly. The Gump header tank is draining, so this caused clogging and low water pressure. This is helped by cleaning the lines and using minimal water flow.
Spawning
We did not observe A. hyacinthus spawning. A. pulchra parents numbers 63, 61, 60, 79, and 65 set and produced bundles that went into the pool for fertilization. Colonies were moved into buckets at 2040 and spawning started at 2100. We collected bundles from colonies and let sit in the bucket to fertilize in the poo. until 2214=5.
Using protocols described on 18 October 2022, we rinsed and allocated 1.5-2 mL of embryos into each squarical numbered C3-6 and D3-6. We also set up 2 extra static squaricals in water baths/water tables as extra material. We also put 1.5-2 mL of embryos into each of 3 5-gallon buckets surrounded by a water bath. We also added embryos into mesh-sided bins (the size of a shoebox) as well as 6 statis tupperware containers. These extra larvae will be used for the Correa group to conduct their larval experiments.
All embryos were added to containers at 0030-0130. High fertilization success was observed at 0240.
18 October 2022
Embryo rearing
Embryos from last night’s spawning were alive and developing this morning at 0630. There was a mixture of fertilized and developing embryos and unfertilized eggs. The unfertilized eggs degraded by 1300. The water quality in the squaricals stayed clean and the systems ran well. Prawn chip stages were observed by 1200.
At 1600 all embryos were removed from the squaricals in preparation for tonight’s spawning. They were poured through a seive and all pooled into one 5 gallon bucket filled with seawater and contained in a water bath in a small blue tank. The bucket was marked with the date of spawning (10/17) and more water was added at 1700 to refresh the culture.
E5 experiment - cleaning tanks
We cleaned all tanks for the E5 experiment today. All fragments look healthy and no mortality was observed.
E5 experiment - loggers
All E5 loggers were downloaded between 1200 and 1400. The logger data was ran using the logger R script in the GitHub repo. Our environmental data looks like this:

Preparing spawning - cleaning tanks
We prepared for spawning by cleaning all coolers, squaricals, tanks, and water lines. We did this twice today due to poor water quality from rain and low water pressure. We will keep a close eye on tanks and lines tonight.
Spawning
Gamete collection for Ariana’s project will follow this spawning protocol adapted from work in Montipora capitata. From last night’s spawning, we learned that A. pulchra bundles break apart quickly, within about 20-30 min from release. Therefore, we will modify this spawning protocol to not do fertilization in the falcon tubes, but rather do fertilization in a 5 gallon bucket in the pool of all collected gametes. After fertilization in this pool, we will use falcon tubes to allocate to squaricals.
The corals started spawning at 2100 with 7 wildtype colonies spawning. Colonies 80, 79, 77, and 61 were individually isolated and sampled for reproductive characteristics by Hollie including eggs per bundle and bundle-bundle crosses. Bundles that were not used for this study were added to bulk fertilization in a bucket pool as described from 17 October 2022 spawning. Bundles from other colonies were collected from the large wildtype tank.
We collected bundles from 2100-2200. The bundles started to break apart after 2120-2130. We prioritized collecting bundles prior to breaking to ensure that we had sperm from each parent in the bucket pool. 7 parents contributed to the pool tonight including 80, 79, 77, 61, 65, 63, and 60. The pool of eggs and sperm were mixed every 10-15 minutes and allowed to sit and fertilized until 2300. The bucket contained a high concentration of sperm (very milky water) and eggs that covered the surface of the bucket.

Fertilized eggs were then added to falcon tubes at 2300-0000 for rinsing and allocating into squaricals. 1.5-2 mL of eggs were added to each falcon tube and allowed to sit so the eggs rose to the top. A seriological pipette was then used to take out as much of the sperm water underneath the eggs as possible. Then 40 mL of 50 um filtered seawater was added as a rinse. Finally, water was removed one more time and the clean embryos were poured into squaricals at 0030. 1 falcon tube was added to each squarical in numbers A1-A6 and B1-B6.

Flow is set at 25-30 mL per 5 seconds in these tanks. Cultures were checked and cleaned every 2-3 hours throughout the night by stirring with a pipette and washing off the banjo filter with the water input hose. Flow was set at 25-30 mL per 5 seconds.
Fertilization was checked at 0215 and we saw 16 and 32 cell stages with low unfertilized eggs, indicates high fertilization success.
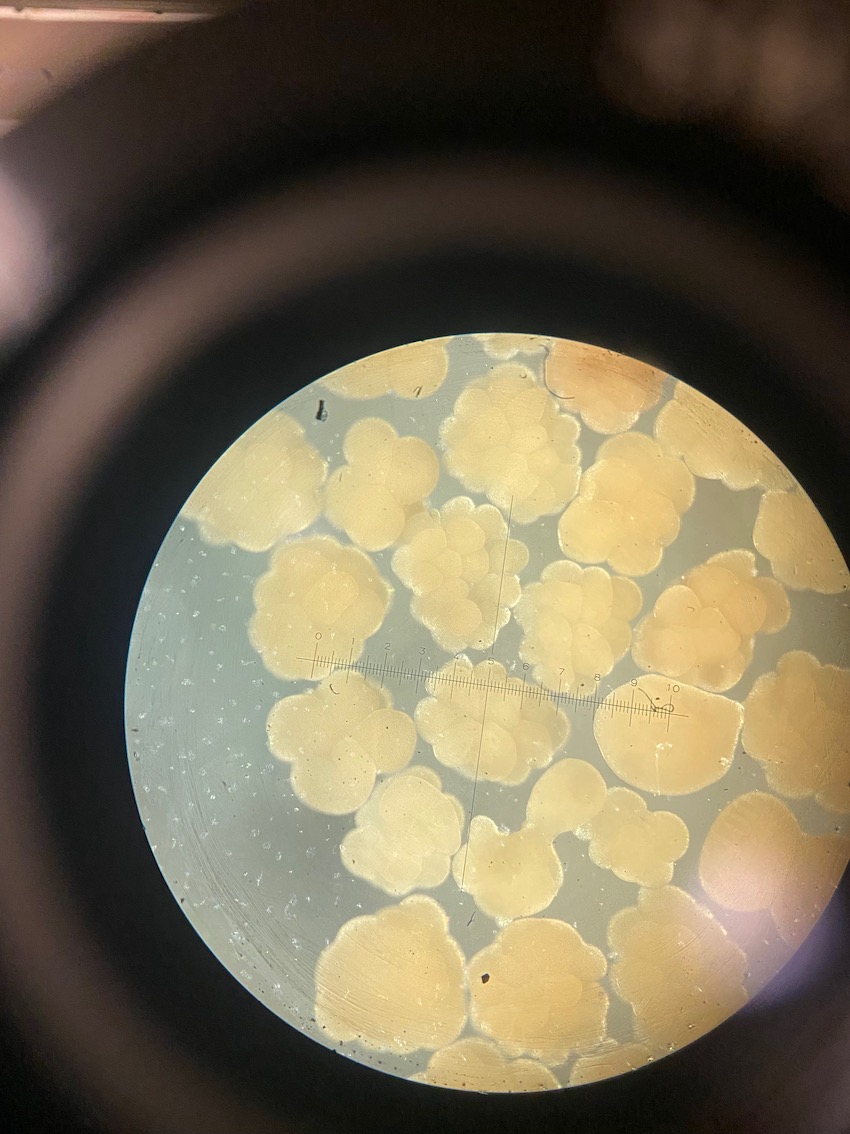
At 0130 we collected 1 fragment per parent colony (#61-80) and froze at -40C in whirlpacks. These will be used for parent molecular samples.
Maintenance of larvae every 2-3 hours includes:
- Checking the water level of the coolers and cleaning the in line 50 um filters
- Using the water line to move embryos off of edges and banjo filter
- Stirring the embryos using a transfer pipette
- Removing any dead material with a pipette
17 October 2022
Daily measurements
Ariana and Terava took daily measurements today between 12:45 and 13:15. Daily measurements are recorded in GitHub here. Tanks will be cleaned tomorrow.
E5 experiment - symbiont counts
Ariana and Terava finished the symbiont density measurements today. The proceedure followed the protocol for E5 processing with no modifications. Data is recorded in GitHub.
Ariana also ran the data using R scripts to analyze data as we go based on the E5 timeseries project scripts.
The symbiont cell density data by colony/genotype looks like this:
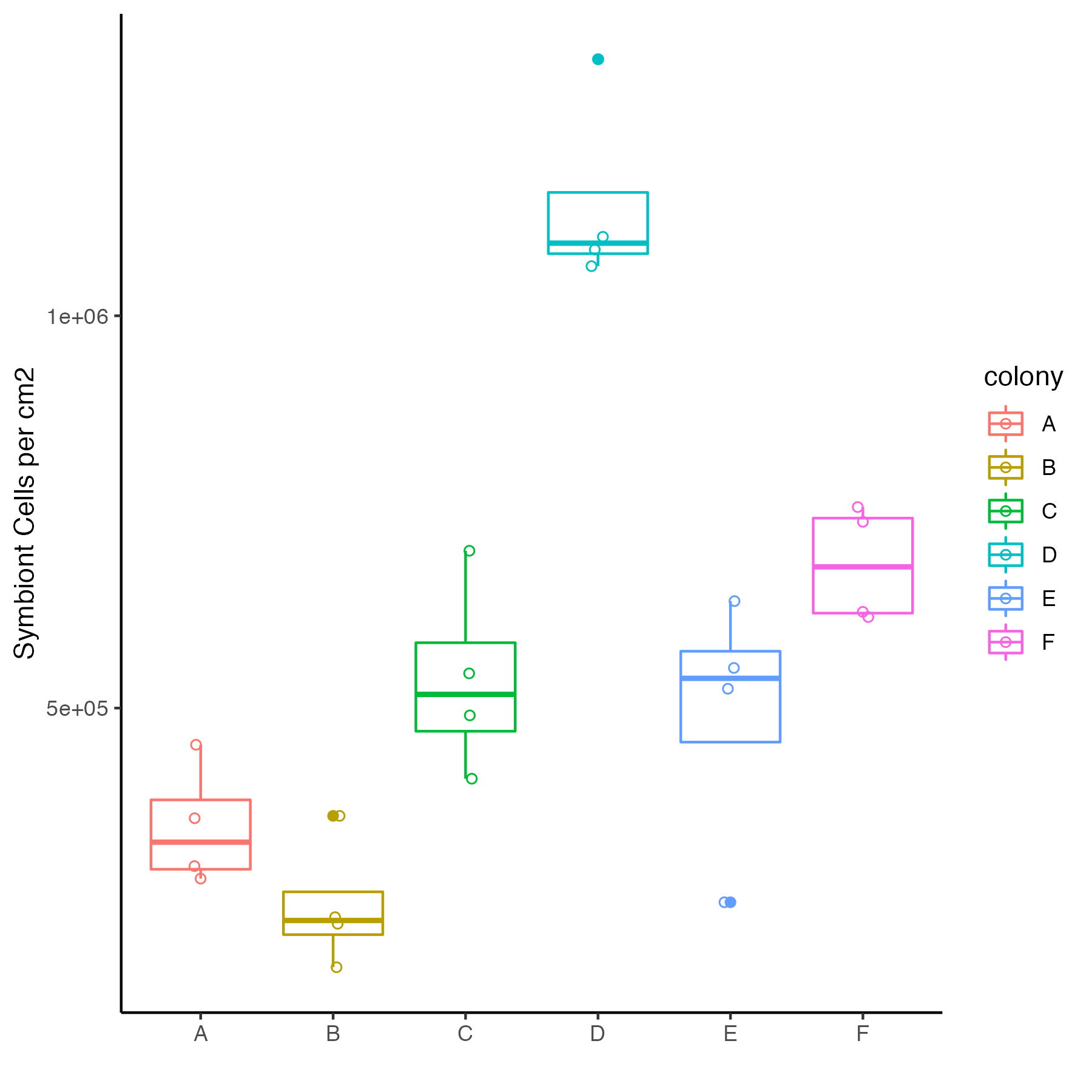
There is a significant difference in symbiont density between colonies.
lmer(cells.cm2~colony + (1|tank), data=.)
p<0.001
E5 experiment - starting treatment
Today we are starting treatments in our E5 A. pulchra metabolism experiment. The treatments for each tank are as follows:
| Tank | Light Treatment | Food Treament |
|---|---|---|
| 1 | Full light | Added food |
| 2 | Full light | No food |
| 3 | Shaded | Added food |
| 4 | Full light | Added food |
| 5 | Shaded | Added food |
| 6 | Full light | No food |
Calculating the amount of food needed
For Coral Max food, the manufacturers instructions suggest 1/2 teaspoon per 50 gallons of tank water. Our tanks are 19.3 gallons (29 cm x 60 cm x 42 cm). Therefore, we would need 0.368 of a 1/2 teaspoon dose (19.3 / 50 gallons).
The mass of 1/2 teaspoon (converted to volume of 2.464 mL) dose is 1.148 g. 1.148 g * 0.368 = 0.422 g. 0.422 g is the amount we need for the size of our tanks. We will round up. For each tank, we will add 0.5 g of food at each feeding.
If we feed 3 times per week in each of 4 tanks (those that have food in the treatment), then we will use 6 g per week. If we run the experiment for 8 weeks, we will need a total of 48 g. We have >200 g, so we have plenty for this experiment.
We then weighed 0.5g into a 5 mL tube and marked the level that the food was at. 0.5g of food is exactly at the 1 mL mark on the white cap 5mL tubes.
We labeled 4 5mL tubes with the tank number for all tanks that recieve food (1, 3, 4, and 5) and marked with a sharpie the 1mL mark to fill food. At each feeding, the person feeding will fill tube to this mark and then add to the tank. See the feeding section below for a protocol on feeding.
E5 experiment - adding shade
We added shade cloth to tanks 3 and 5 at 13:09 with one layer of shade cloth. We recorded light in the tanks after shade cloth was added. Daily measurements were taken before shade cloth was added. The values of light are included below that show the comparison of light before and after shading in each tank.
Light before adding shade:
| Tank | Time | Center | Q1 | Q2 | Q3 | Q4 |
|---|---|---|---|---|---|---|
| 3 | 1249 | 96 | 120 | 118 | 116 | 115 |
| 5 | 1252 | 116 | 103 | 102 | 83 | 90 |
Light after adding shade:
| Tank | Time | Center | Q1 | Q2 | Q3 | Q4 |
|---|---|---|---|---|---|---|
| 3 | 1309 | 59 | 65 | 56 | 65 | 53 |
| 5 | 1310 | 38 | 36 | 31 | 38 | 32 |
Light is reduced by approx. 35-55% after adding shade.

E5 experiment - feeding
The process for feeding is as follows and is described in the E5 A. pulchra metabolism feeding protocol here. The overview of the steps are as follows:
- Measure out food as described above into labeled 5 mL tubes to the marked 1 mL mark with Coral Max.
- Turn off water on the main line to all tanks.
- Use the nalgene bottle or a tri pour to take ~ 1 L water out of the tanks to ensure the food doesn’t overflow.
- Add the food to each respective tank (tanks 1, 3, 4, 5) and stir and break up the chunks with a transfer pipette.
- Start a timer for 1 h.
- After 1 h, turn the water flow back on.
Repeat this process 3 times per week (e.g., Mon, Wed, Fri).
Record the timing and notes about feeding in the schedule tracking document here.
Today coral feeding started at 13:53 and water was turned on at 14:55.
Spawning
We watched for spawning in A. hyacinthus and A. pulchra. We observed several of our A. pulchra colonies spawning tonight (3-4 wildtype and 5-6 of Danielle’s heatwave colonies). Spawning started at 21:10. Bundles broke apart very quickly - we did not observe many bundles in the wildtype collection, they rapidly broke apart and we collected eggs from the tank. We did not notice any obvious setting in the colonies before hand.
Interestingly, several fragments from the E5 tank experiment showed setting and spawning!
We added ~1-2 mL of bundles/eggs to n=6 falcon tubes. We had a limited amount of material. Gametes were scooped from the water using gravy separators. The separators were then allowed to sit for 1-2 min to allow the bundles to come up to the surface. Water was then drained from the bottom and the gametes were mixed together in a 5 gallon bucket for the pooled wildtype fertilization.
Gametes were finished being added to falcon tubes at 00:30. This took longer than we would have liked as the amount of material was low and it took a long time to individually pipette embryos to try to get as much material as possible.
During this time in the large tank, the embryos fertilized. At 13:00 we observed 2-cell and 4-cell stages under the microscope. After this, Ariana added one of each of the falcon tubes to n=6 squaricals. These were added to the squarials at 02:00. These are low density and we will check on them regularly.

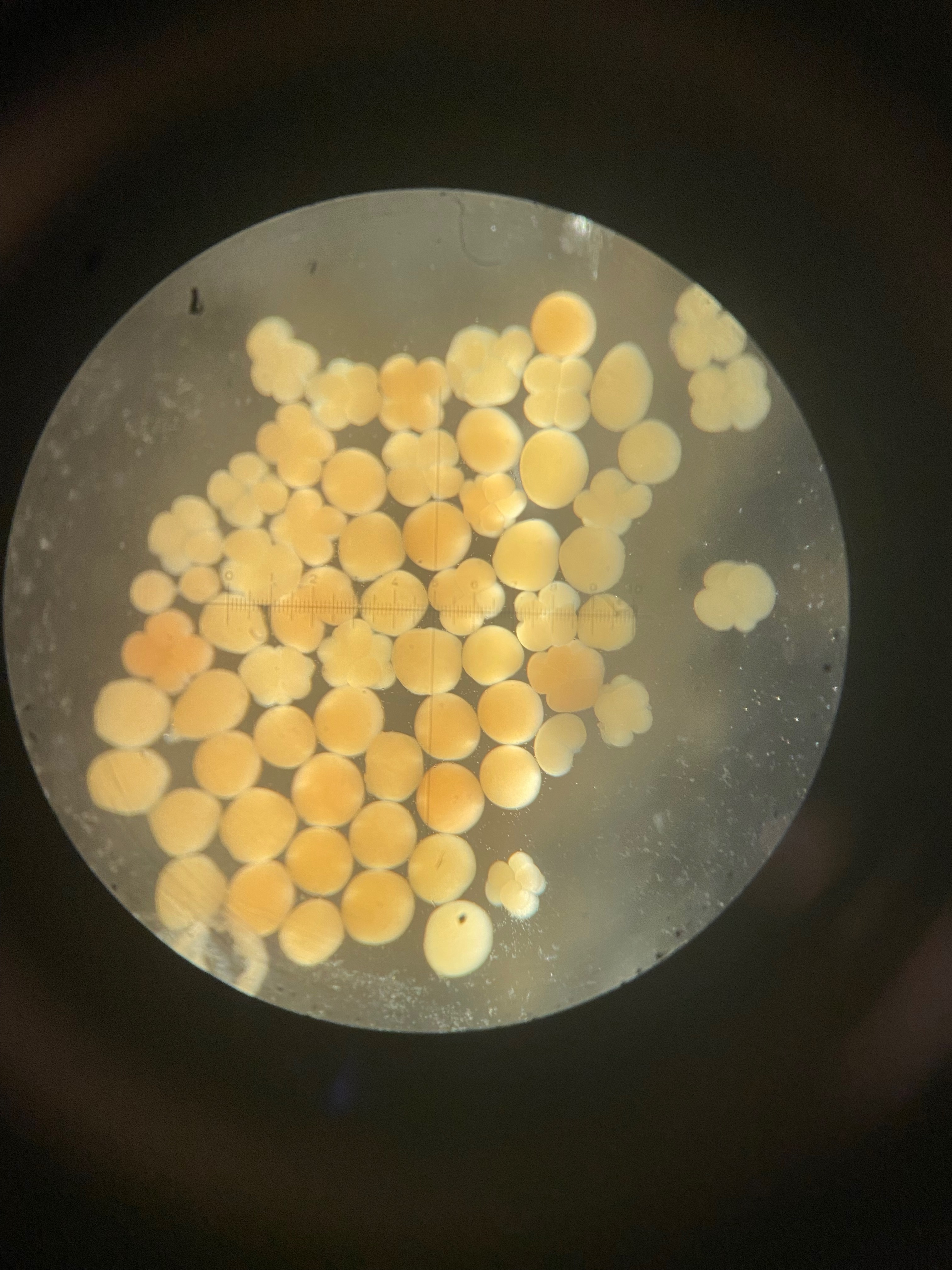

Danielles colonies:
16 October 2022
Hollie arrived today and joined our field team!
Daily measurements
Ariana and Danielle took daily measurements in the E5 experimental tanks as well as the tanks holding adult colonies between 1345 and 1415. Daily measurements are recorded in GitHub here.
Tank cleaning
All coolers and squaricals were cleaned and rinsed with fresh water today. The system was restarted before we prepared for spawning tonight.
Spawning
We watched for spawning in A. hyacinthus and A. pulchra from 2000-2330. No spawning was observed.
15 October 2022
Daily measurements
Ariana took daily measurements in the E5 experimental tanks as well as the tanks holding adult colonies between 1400 and 1430. Daily measurements are recorded in GitHub here.
October 2022 Field Day Sampling - Backreef Acropora and Pocillopora
Danielle, Ariana, and Pierrick conducted monthly timepoint sampling at the Mahana field site (17°29’12.5”S 149°53’16.1”W). Following the Field Sampling Protocol, we collected 40 ~5 inch Acropora pulchra fragments from the five patches located on the site. Additionally, 10 4-5 inch fragments of Pocillopora were collected and brought back to Gump Station as well for same day processing. For all sampling locations, a close up photo with the tag was taken. Additionally, a bleaching score photo was taken at each sampling location. If a tag was not found, we sampled from the same colony that the tag should have been located on.
After collecting the samples, Danielle followed the Same Day Sample Processing Protocol to prepare samples for histological analysis, molecular and physiological downstream processing.
All bleaching score photos were downloaded off of the Putnam Lab Canon G7X camera number 2 and uploaded to Pocillopora_Sampling_Colony_Photos and Acropora_Sampling_Colony_Photos for the October 2022 timepoint.
All timepoint and tracking data sheets for the October 2022 timepoint for both Pocillopora and Acropora were updated.
E5 experiment - symbiont counts
Ariana counted symbiont density in more samples today. The proceedure followed the protocol for E5 processing with no modifications. Data is recorded in GitHub.
Ariana also wrote scripts to analyze data as we go based on the E5 timeseries project scripts.
Preliminary data shows that symbiont densities (~1e+06) are in the range seen in the E5 timeseries project.

The remaining samples will be processed over the next few days.
Spawning
We watched for spawning in A. hyacinthus and A. pulchra from 2000-2330. No spawning was observed.
Tank cleaning
We cleaned all larval coolers and flushed all water lines to improve water flow at 2100.
14 October 2022
A. pulchra high frequency sampling dissections
Danielle finished processing the A. pulchra samples from the sixth high frequency sampling (20221012) for dissections. The fragments from this timepoint had many more distinct gonads.
Daily measurements
Ariana and Teravatook daily measurements in the E5 experimental tanks as well as the tanks holding adult colonies between 1000 and 1100. Daily measurements are recorded in GitHub here.
E5 experiment - symbiont counts
Ariana and Terava started symbiont cell density measurements on the symbiont fraction of samples that were separated yesterday. The proceedure followed the protocol for E5 processing with no modifications.
Data is recorded in GitHub. Five samples were analyzed today. The remaining samples will be processed over the next few days.
E5 experiment - analyzing data
Ariana wrote scripts to analyze protein and surface area for the baseline E5 adult measurements. The scripts are located on GitHub.
Protein measurements from yesterday were briefly analyzed. These are the protein values for each colony/genotype in the host and holobiont fraction in the figure below.
A linear mixed model testing differences in protein by genotype show a significant difference in both host and holobiont protein by genotype.
lmer(protein_ug.cm2~colony + (1|tank), data=protein)
Labeling adult colonies
Adult colonies were labeled with orange cattle tags attached with zip ties. A. pulchra tags were no. 061-080. A. hyacinthus tags were no. 081-096.
Spawning
Ariana prepared tubes labeled no. 1-96 (egg and sperm samples)and prepared 24-well plates (eggs per bundle) and scintillation vials (fertilization success) for potential spawning for individual characterization of wild colonies.
No spawning was observed in either species tonight.
13 October 2022
Daily measurements
Ariana took daily measurements in the E5 experimental tanks as well as the tanks holding adult colonies between 1030 and 1100. Daily measurements are recorded in GitHub here.
E5 experiment - total protein
Ariana, Danielle, and Terava conducted total soluble protein assays (Pierce BCA protein assay) for host and holobiont fractions for Danielle’s histology time series samples and the E5 adult baseline samples following the E5 total protein protocol.
The summary of the protocol process for Ariana’s samples was as follows. Danielle and Ariana followed the same protocol with an added step for host-symbiont fraction separation for Ariana’s samples. 2 plates were run for host samples and 2 plates were run for holobiont samples in total.
- Thaw two 1mL tube samples for each E5 fragment.
- For one of the tubes, we separated host and symbiont fraction. The second tube will have the holobiont fraction.
- One tube per fragment was thawed and centrifuged at 3500 rpm for 3 min using the Fisher centrifuge as used during symbiont isolation trials on 3 Oct 2022.
- The supernatant (host fraction) was transfered to a new tube labeled with the fragment ID and “H” for “host”. The tube with the symbiont pellet was labeled with an “S” for “symbiont”.
- For each sample, this resulted in one tube with holobiont tissue, one tube with host tissue, and one tube with symbiont pellet.
- The symbiont fraction was returned to the freezer at -40C for symbiont counting tomorrow.
- Standards and working solution were generated as described in the total protein protocol.
- First, host fraction samples were run by loading 25 uL of each sample in duplicates in a 96 well plate along with duplicates of each standard. The plate map for host samples is below.
- The plate was gently rocked by hand to mix for 30 sec and then incubated at 37°C for 30 minutes.
- After incubation, the plate was allowed to cool at room temperature for 6 min.
- The absorbance of the plate was read on the microplate spectophotometer reader at 562 nm and saved.
- In the next plate, holobiont samples were added in duplicates with 25 uL of each sample and each standard. The plate map is included below.
- The plate was incubated and read in the same way as described above.
- All plates and supplies were cleaned following plate reading.
Host samples were added to the incubator at 1313 and read at 1351. Holobiont samples were added to the incubator at 1356 and read at 1430.
A host and holobiont plate was run prior to these samples by Danielle for the gametogenesis time series.
The data was saved and added to E5 GitHub here and the Gametogenesis Github here.
Plate Map Host (September Gametogenesis 2022)
| well | colony_id | |||
|---|---|---|---|---|
| A1 | StandardA | |||
| A2 | StandardA | |||
| A3 | StandardB | |||
| A4 | StandardB | |||
| A5 | StandardC | |||
| A6 | StandardC | |||
| A7 | StandardD | |||
| A8 | StandardD | |||
| A9 | StandardE | |||
| A10 | StandardE | |||
| A11 | StandardF | |||
| A12 | StandardF | |||
| B1 | StandardG | |||
| B2 | StandardG | |||
| B3 | StandardH | |||
| B4 | StandardH | |||
| B5 | StandardI | |||
| B6 | StandardI | |||
| B7 | ACR-414 | |||
| B8 | ACR-414 | |||
| B9 | ACR-415 | |||
| B10 | ACR-415 | |||
| B11 | ACR-416 | |||
| B12 | ACR-416 | |||
| C1 | ACR-417 | |||
| C2 | ACR-417 | |||
| C3 | ACR-418 | |||
| C4 | ACR-418 | |||
| C5 | ACR-420 | |||
| C6 | ACR-420 | |||
| C7 | ACR-421 | |||
| C8 | ACR-421 | |||
| C9 | ACR-422 | |||
| C10 | ACR-422 | |||
| C11 | ACR-423 | |||
| C12 | ACR-423 | |||
| D1 | ACR-424 | |||
| D2 | ACR-424 | |||
| D3 | ACR-425 | |||
| D4 | ACR-425 | |||
| D5 | ACR-426 | |||
| D6 | ACR-426 | |||
| D7 | ACR-427 | |||
| D8 | ACR-427 | |||
| D9 | ACR-428 | |||
| D10 | ACR-428 | |||
| D11 | ACR-429 | |||
| D12 | ACR-429 | |||
| E1 | ACR-430 | |||
| E2 | ACR-430 | |||
| E3 | ACR-431 | |||
| E4 | ACR-431 | |||
| E5 | ACR-432 | |||
| E6 | ACR-432 | |||
| E7 | ACR-433 | |||
| E8 | ACR-433 | |||
| E9 | ACR-434 | |||
| E10 | ACR-434 | |||
| E11 | ACR-435 | |||
| E12 | ACR-435 | |||
| F1 | ACR-436 | |||
| F2 | ACR-436 | |||
| F3 | ACR-437 | |||
| F4 | ACR-437 | |||
| F5 | ACR-438 | |||
| F6 | ACR-438 | |||
| F7 | ACR-439 | |||
| F8 | ACR-439 | |||
| F9 | ACR-446 | |||
| F10 | ACR-446 | |||
| F11 | ACR-447 | |||
| F12 | ACR-447 | |||
| G1 | ACR-448 | |||
| G2 | ACR-448 | |||
| G3 | ACR-449 | |||
| G4 | ACR-449 | |||
| G5 | ACR-451 | |||
| G6 | ACR-451 | |||
| G7 | ACR-456 | |||
| G8 | ACR-456 | |||
| G9 | ACR-457 | |||
| G10 | ACR-457 | |||
| G11 | ACR-458 | |||
| G12 | ACR-458 | |||
| H1 | ACR-459 | |||
| H2 | ACR-459 | |||
| H3 | ACR-460 | |||
| H4 | ACR-460 | |||
| H5 | ACR-461 | |||
| H6 | ACR-461 | |||
| H7 | ACR-462 | |||
| H8 | ACR-462 | |||
| H9 | ACR-463 | |||
| H10 | ACR-463 | |||
| H11 | ACR-464 | |||
| H12 | ACR-464 |
Plate Map Holobiont (September Gametogenesis 2022):
| well | colony_id | |||
|---|---|---|---|---|
| A1 | StandardA | |||
| A2 | StandardA | |||
| A3 | StandardB | |||
| A4 | StandardB | |||
| A5 | StandardC | |||
| A6 | StandardC | |||
| A7 | StandardD | |||
| A8 | StandardD | |||
| A9 | StandardE | |||
| A10 | StandardE | |||
| A11 | StandardF | |||
| A12 | StandardF | |||
| B1 | StandardG | |||
| B2 | StandardG | |||
| B3 | StandardH | |||
| B4 | StandardH | |||
| B5 | StandardI | |||
| B6 | StandardI | |||
| B7 | ACR-414 | |||
| B8 | ACR-414 | |||
| B9 | ACR-415 | |||
| B10 | ACR-415 | |||
| B11 | ACR-416 | |||
| B12 | ACR-416 | |||
| C1 | ACR-417 | |||
| C2 | ACR-417 | |||
| C3 | ACR-418 | |||
| C4 | ACR-418 | |||
| C5 | ACR-420 | |||
| C6 | ACR-420 | |||
| C7 | ACR-421 | |||
| C8 | ACR-421 | |||
| C9 | ACR-422 | |||
| C10 | ACR-422 | |||
| C11 | ACR-423 | |||
| C12 | ACR-423 | |||
| D1 | ACR-424 | |||
| D2 | ACR-424 | |||
| D3 | ACR-425 | |||
| D4 | ACR-425 | |||
| D5 | ACR-426 | |||
| D6 | ACR-426 | |||
| D7 | ACR-427 | |||
| D8 | ACR-427 | |||
| D9 | ACR-428 | |||
| D10 | ACR-428 | |||
| D11 | ACR-429 | |||
| D12 | ACR-429 | |||
| E1 | ACR-430 | |||
| E2 | ACR-430 | |||
| E3 | ACR-431 | |||
| E4 | ACR-431 | |||
| E5 | ACR-432 | |||
| E6 | ACR-432 | |||
| E7 | ACR-433 | |||
| E8 | ACR-433 | |||
| E9 | ACR-434 | |||
| E10 | ACR-434 | |||
| E11 | ACR-435 | |||
| E12 | ACR-435 | |||
| F1 | ACR-436 | |||
| F2 | ACR-436 | |||
| F3 | ACR-437 | |||
| F4 | ACR-437 | |||
| F5 | ACR-438 | |||
| F6 | ACR-438 | |||
| F7 | ACR-439 | |||
| F8 | ACR-439 | |||
| F9 | ACR-446 | |||
| F10 | ACR-446 | |||
| F11 | ACR-447 | |||
| F12 | ACR-447 | |||
| G1 | ACR-448 | |||
| G2 | ACR-448 | |||
| G3 | ACR-449 | |||
| G4 | ACR-449 | |||
| G5 | ACR-451 | |||
| G6 | ACR-451 | |||
| G7 | ACR-456 | |||
| G8 | ACR-456 | |||
| G9 | ACR-457 | |||
| G10 | ACR-457 | |||
| G11 | ACR-458 | |||
| G12 | ACR-458 | |||
| H1 | ACR-459 | |||
| H2 | ACR-459 | |||
| H3 | ACR-460 | |||
| H4 | ACR-460 | |||
| H5 | ACR-461 | |||
| H6 | ACR-461 | |||
| H7 | ACR-462 | |||
| H8 | ACR-462 | |||
| H9 | ACR-463 | |||
| H10 | ACR-463 | |||
| H11 | ACR-464 | |||
| H12 | ACR-464 |
Plate Map Host (E5 samples):
| Row | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | STND-A | STND-A | STND-B | STND-B | STND-C | STND-C | STND-D | STND-D | STND-E | STND-E | STND-F | STND-F |
| B | STND-G | STND-G | STND-H | STND-H | STND-I | STND-I | A7 | A7 | A16 | A16 | A20 | A20 |
| C | A31 | A31 | B9 | B9 | B20 | B20 | B25 | B25 | B34 | B34 | C1 | C1 |
| D | C19 | C19 | C26 | C26 | C30 | C30 | D4 | D4 | D13 | D13 | D26 | D26 |
| E | D30 | D30 | E5 | E5 | E8 | E8 | E20 | E20 | E31 | E31 | F2 | F2 |
| F | F9 | F9 | F17 | F17 | F22 | F22 | ACR465 | ACR465 | - | - | - | - |
| G | - | - | - | - | - | - | - | - | - | - | - | - |
| H | - | - | - | - | - | - | - | - | - | - | - | - |
Plate Map Holobiont (E5 samples):
| Row | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | STND-A | STND-A | STND-B | STND-B | STND-C | STND-C | STND-D | STND-D | STND-E | STND-E | STND-F | STND-F |
| B | STND-G | STND-G | STND-H | STND-H | STND-I | STND-I | A7 | A7 | A16 | A16 | A20 | A20 |
| C | A31 | A31 | B9 | B9 | B20 | B20 | B25 | B25 | B34 | B34 | C1 | C1 |
| D | C19 | C19 | C26 | C26 | C30 | C30 | D4 | D4 | D13 | D13 | D26 | D26 |
| E | D30 | D30 | E5 | E5 | E8 | E8 | E20 | E20 | E31 | E31 | F2 | F2 |
| F | F9 | F9 | F17 | F17 | F22 | F22 | ACR465 | ACR465 | - | - | - | - |
| G | - | - | - | - | - | - | - | - | - | - | - | - |
| H | - | - | - | - | - | - | - | - | - | - | - | - |
Spawning
We started water in the spawning systems at 1900 and prepared equipment for possible spawning. We did not observe any setting or spawning in either A. hyacinthus or A. pulchra.
12 October 2022
Collecting parent colonies
Danielle and Ariana spent the day collecting parent colonies in preparation for spawning. The following coral colonies were collected:
- 24 experimental heatwave A. pulchra colonies for Danielle’s project collected from Mahana from 0900-1100.
- 20 wildtype A. pulchra colonies for Ariana’s project and collaborative projects collected from Mahana from 1100-1200.
- 16 wildtype A. hyacinthus colonies for Ariana’s project and collaborative projects collected from Manava near the E5 site and Pierrick’s tagged Pocillopora site from 1200-1330.
Wildtype colonies were sampled using a hammer and chisel. A. hyacinthus colonies had a tendency to break in to multiple pieces, these were placed next to each other in the tanks. Collections were across a range of colony sizes from different patches/locations within each species.
Colonies were placed in n=2 tanks for Danielle’s colonies with n=1 tank for each wildtype species. Water flow was kept very high with n=3 recirculating pumps in each tank.
A. pulchra high frequency sampling - 20221012
Completed the sixth timepoint for the high frequency project for A. pulchra on the samples below. Went in the field at 15:00 to collect 20 fragments from the five patches.
Metadata:
| Species | Site | OrderbyPatch | Original.Tag.Num | Patch | Latitude.S | Longitude.W | GPS.Point.Name | Timestamp | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. pulchra | Mahana | P1.3 | 414 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:59 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.5 | 416 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:11 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.4 | 424 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:17 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.1 | 426 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:49 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.1 | 429 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:05 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.4 | 430 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:10 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.9 | 431 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:46 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.5 | 433 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:02 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.4 | 434 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.10 | 436 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:29 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.2 | 437 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:50 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.10 | 439 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:47 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.1 | 448 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:35 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.2 | 449 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:07 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.8 | 451 | P5 | 17°29.220’ | 149°53.236’ | P5 | 2:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.6 | 458 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.11 | 461 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:30 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.1 | 462 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.6 | 464 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:41 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.6 | 465 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:22 PM |
A. pulchra high frequency sampling protocol
Danielle followed the same day sample processing protocol for the 20 collected high frequency samples. Danielle also set aside 10 fragments to be dissected and preserved in DNA/RNA shield.
Spawning preparations
Danielle prepared all the spawning supplies required for her project using the same set up and workflow that was used in September.
Ariana set up all squaricals, coolers, and Apex systems as described for the September spawning. All systems ran smoothly today with sufficient water flow.
The Putnam Lab and Correa Lab teams met together to talk about coral spawning and do an orientation to the current workflows and tank set up.
Ariana repaired mesh on plastic tupperware containers that will be extra containers to rear larvae if we have an excess of spawning material.
Loggers
Ariana launched Hobo at 0900, logging every 10 min in the parent tanks prior to colony collection:
- Wildtype parent 2 SSN 20719656
- DBP Parent 2 SSN 20444040
- DBP Parent 1 SSN 20946644
- Wildtype parent 1 SSN 20444033
Hobo pendants for the larval containers were also launched at 2100 logging every 10 min. Loggers were launched for A1-3, B1-3, C1-3, and D1-3 tanks with the same SSN assignments as on September 16 2022.
11 October 2022
E5 adult coral metabolism project
Daily measurements
Ariana and Danielle took daily measurements of E5 tanks between 1500 and 1530. No mortality was observed. Daily measurements are recorded in GitHub here. Ariana analyzed the daily measurement data in this R script. Total pH was calculating using the Tris calibration from 09/24/22 following the Silbiger Lab protocol and scripts. The data looks like this:
We used one-way ANOVA tests to test for differences in each of these metrics between tanks. There were no significant differences detected.
New coral tags
Ariana continued replacing coral tags for the E5 tanks. All tags were replaced.
Tank cleaning
Ariana and Danielle cleaned all E5 tanks and all corals. Tanks 1, 2, and 3 were moved to a water table further under the shade structure. In the mornings, tanks 1, 2, and 3 have been getting intense morning light at sunrise. Moving the tanks further under the building will make the light more even.

Loggers
Ariana downloaded all Hobo pendant loggers and conducted a preliminary analysis of logger data. We saw that tanks 1, 2, and 3 were getting higher light and temperature peaks in the morning just after sunrise due to the angle of the sun on these tanks. Because of this, we rearranged the tanks to move them under the building structure as described above. The logger data for the E5 tanks is shown below:
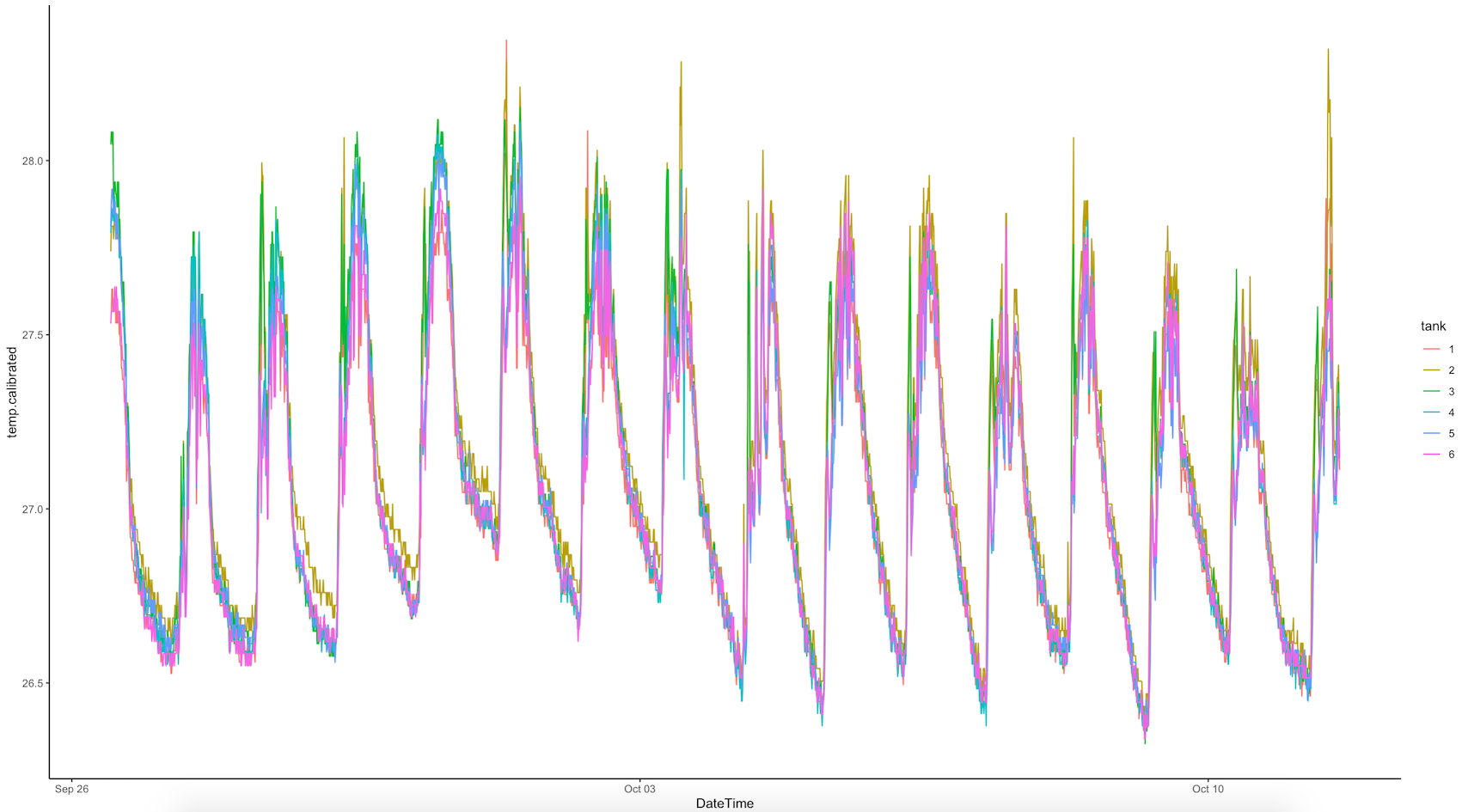
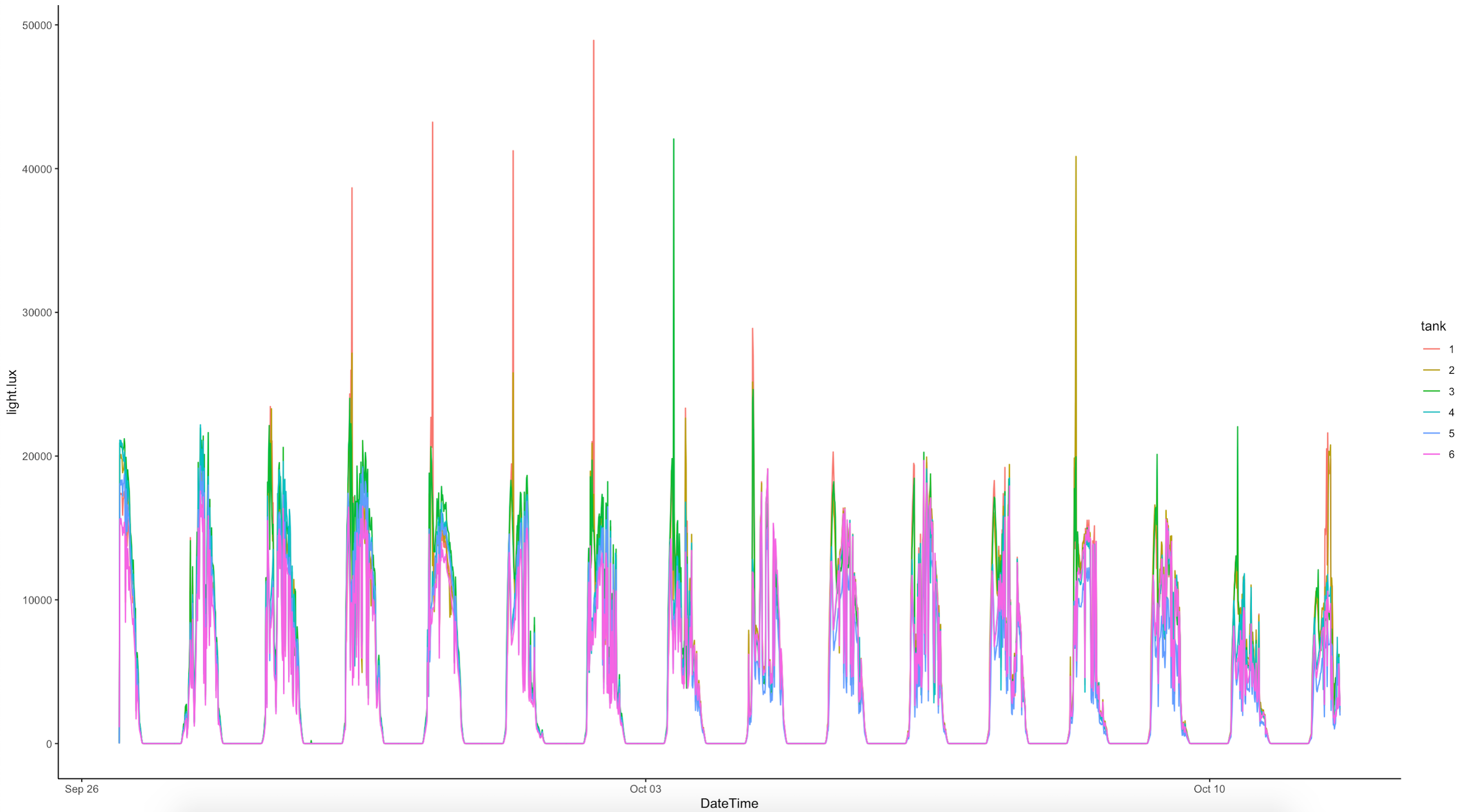
Light from the Odyssey logger is reading in Integrated PAR, which will be higher than instantaneous PAR recorded by the Apogee sensor for daily measurements.
We also plotted the temperatures from the heatwave and wildtype A. pulchra parent colonies.
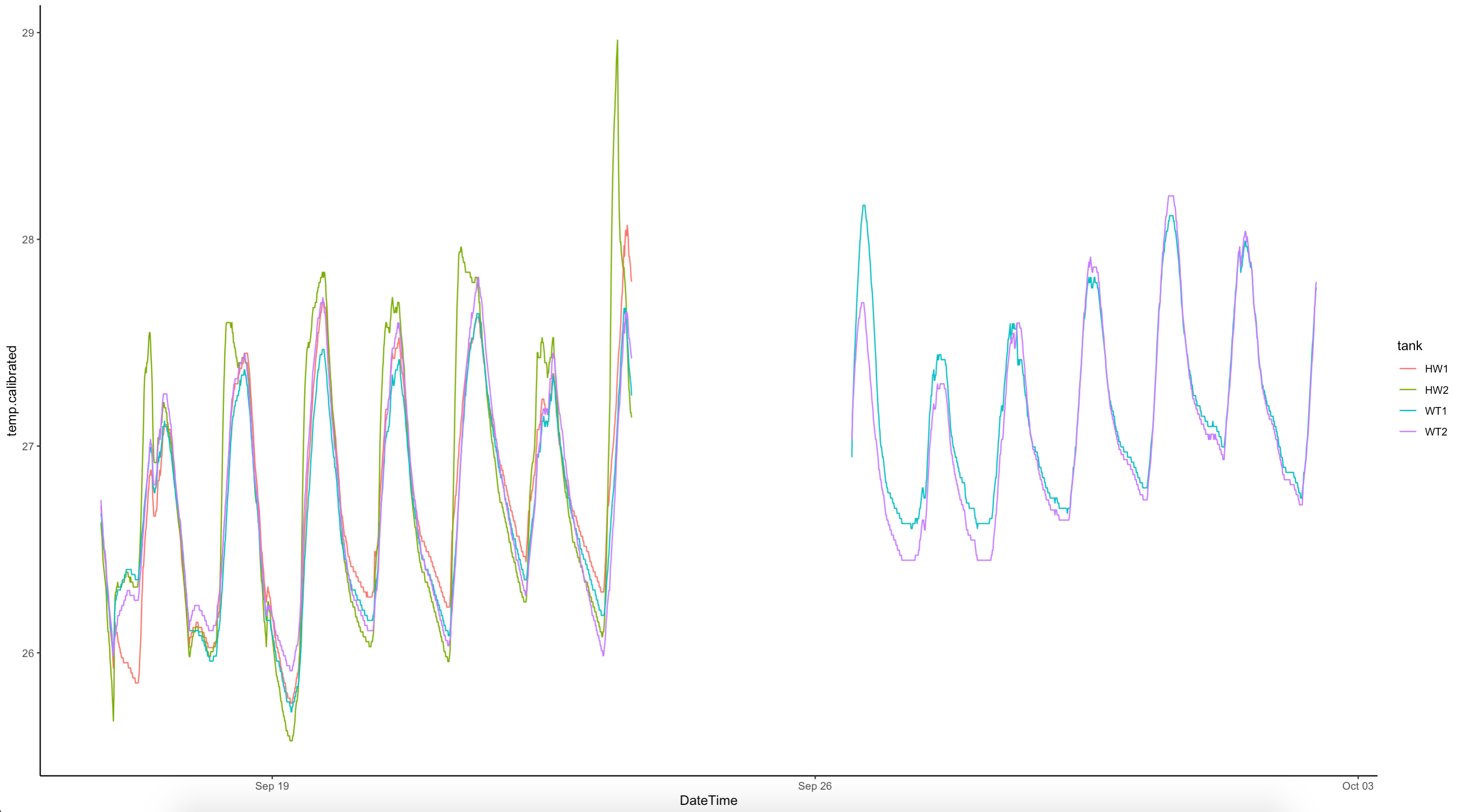
Spawning preparations
Ariana and Danielle cleaned and set up the coolers and squaricals in preparation for another round of spawning in the next week. We also filled and cleaned the large blue tanks in preparation for collecting adult colonies tomorrow from the field.

10 October 2022
Today Terava Atger began working with us supported by Danielle’s NSF GRFP funding. Terava will work with us on all projects over the next 3 weeks including the E5 A. pulchra metabolism experiment, histology time series, and coral spawning.
E5 adult coral metabolism project
Daily measurements
Ariana and Teravatook daily measurements of E5 tanks between 1000 and 1045. No mortality was observed. Light values were variable during measurements due to passing clouds. Daily measurements are recorded in GitHub here.
New coral tags
Ariana continued replacing coral tags for the E5 tanks.
Aliquoting tissue
Ariana and Terava aliquoted coral tissue slurries for future analysis of symbiont cell counts and total protein. This was completed between 1300-1430. This is the process we followed:
- Thaw a subset of 50mL falcon tubes with frozen tissue slurry, keeping slurry on frozen beads to thaw slowly.
- All homogenate transferred to a labeled 15mL tube. This is used due to low slurry volume in order to homogenize slurry. All 50mL falcon tubes will be cleaned and re used.
- Tissue homogenized for 10-12 sec with the ProScientific PRO200 microtip homogenizer.
- 4 aliquots of 1 mL slurry were added into n=4 snap cap tubes labeled with the fragment ID. In cases where there was not enough to fill all 4 tubes, volumes were noted on data sheet.
- Aliquots were frozen at -40C in a box labeled with “E5 A. pulchra metabolism; Putnam; aliquot tissue slurry; N=24 fragments; n=4 aliquots per fragment”. There are two boxes (Box #3, Box #4).
- This process was repeated for N=24 fragment slurries, with n=96 total tubes with 1 mL aliquots. All remaining slurry in 15 mL tubes was kept in black mesh bag at -40C.
- All equipment was cleaned and sterilized.
The sample tracking sheet can be found here with information on aliquoting sample locations.
A. pulchra high frequency sampling dissections
Danielle finished processing the A. pulchra samples from the fifth high frequency sampling (20221007) for dissections. The fragments from this timepoint had many more distinct gonads.
9 October 2022
E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements of E5 tanks between 1600 and 1630. No mortality was observed. Light values were low in the afternoon due to cloud cover. Daily measurements are recorded in GitHub here.
New coral tags
Ariana started replacing paper clips with coated wire tags. These will be replaced over the course of the next week.
Pocillopora spawning observation
The team snorkeled at Manava (Ariana and Pierrick) and Hilton (Danielle and Noam) from 0800-1000. Another researcher potentially observed Pocillopora spawning at around 9am at the Hilton site. We decided to snorkel at this later time to see if spawning would occur. We did not see evidence of spawning today.
Histology time series sampling
Danielle and the team sampled a subset of the high frequency time series A. pulchra colonies today for Danielle to do gamete dissections.
7 October 2022
A. pulchra high frequency sampling - 20221007
Completed the fifth timepoint for the high frequency project for A. pulchra on the samples below. Went in the field at 09:00 to collect 20 fragments from the five patches.
Metadata:
| Species | Site | OrderbyPatch | Original.Tag.Num | Patch | Latitude.S | Longitude.W | GPS.Point.Name | Timestamp | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. pulchra | Mahana | P1.3 | 414 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:59 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.5 | 416 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:11 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.4 | 424 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:17 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.1 | 426 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:49 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.1 | 429 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:05 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.4 | 430 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:10 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.9 | 431 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:46 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.5 | 433 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:02 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.4 | 434 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.10 | 436 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:29 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.2 | 437 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:50 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.10 | 439 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:47 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.1 | 448 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:35 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.2 | 449 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:07 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.8 | 451 | P5 | 17°29.220’ | 149°53.236’ | P5 | 2:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.6 | 458 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.11 | 461 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:30 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.1 | 462 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.6 | 464 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:41 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.6 | 465 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:22 PM |
A. pulchra high frequency sampling protocol
Danielle followed the same day sample processing protocol for the 20 collected high frequency samples. Danielle also set aside 10 fragments to be dissected and preserved in DNA/RNA shield.
E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements of E5 tanks between 1400 and 1430. No mortality was observed. Daily measurements are recorded in GitHub here.
Surface area measurements
Ariana measured surface area using the wax dipping method on all E5 baseline fragments. All airbrushed fragments from 5 October 2022 were dried in a drying oven at 80°C for 6 hours. The fragments were then weighed on a Mettler Toledo Excellence Plus SP203S scale to 0.001 g. This was recorded as the fragment dry weight. Fragments were then dipped once in melted wax and allowed to dry. Fragments were then re-weighed on the scale and recorded as wax dipped weight.
We will use the standard curve for this wax batch from the one generated by Danielle during this field trip. The data for the curve is here and the script for processing is here.
Ariana will next work on the scripts to process surface area data. Symbiont cell density and total protein will be the next steps in processing fragments.
Surface area data was recorded in GitHub.
Pocillopora population transects
Today the team completed the last (7 out of 7 completed) transect at the Manava site for Pierrick Harnay’s PhD project on the genetic population of Pocillopora spp. corals in Mo’orea. More information can be found in his notebook here.
Pocillopora spawning observation
The team snorkeled at Manava from 0600-0830 this morning to look for Pocillopora spawning. No spawning was observed.
6 October 2022
E5 adult coral metabolism project
Daily measurements
Ariana and Danielle took daily measurements of E5 tanks between 1000 and 1030. No mortality was observed. Daily measurements are recorded in GitHub here.
Pocillopora population transects
Today the team completed 3 more (6 out of 7 completed) transects at the Manava site for Pierrick Harnay’s PhD project on the genetic population of Pocillopora spp. corals in Mo’orea. More information can be found in his notebook here. The last transect will be completed tomorrow morning during early morning observation for Pocillopora spawning.
5 October 2022
A. pulchra high frequency sampling dissections
Danielle finished processing the A. pulchra samples from the fifth high frequency sampling (20221009) for dissections. The fragments from this timepoint had many more distinct gonads.
E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements of E5 tanks between 1200 and 1230. No mortality was observed. Daily measurements are recorded in GitHub here. Ariana made more coated wire clips and will work on replacing these later this week.
Processing baseline fragments
Fragments that were airbrushed yesterday were removed from 10% bleach cups. Skeletons were clean and placed back in whirl pack bags. These will be processed for wax dipping to measure surface area. We will use the standard curve for this wax batch from the one generated by Danielle during this field trip. The data for the curve is here and the script for processing is here.
Pocillopora population transects
Today the team completed transects at the Manava site for Pierrick Harnay’s PhD project on the genetic population of Pocillopora spp. corals in Mo’orea. More information can be found in his notebook here.
4 October 2022
Meeting with Terava for collaboration and outreach initiatives
Danielle and Ariana met with Terava Atger about collaborations and helping out during the month of October for spawning, physiological analyses, future outreach work partnering with the ‘Ātiti’a center, and future collaboration for data analyses and writing. Terava is enthusiastic about learning new laboratory and research skills as she has been working with Lily Zhao of the Stier group from UCSB and is looking to start her masters or PhD work in the future. See below for general meeting notes:
- We described the various types of experiments we will be conducting from now until the last week in October which include various aspects of field and lab work. coral reproduction and spawning coral monthly reproduction time series coral monitoring in the lagoon coral feeding/metabolism experiment coral symbiosis with algal endosymbionts after spawning
- We discussed your main interests in these projects and what you would like to take away and learn, along with your general availability this month.
- We also discussed the development of ‘Ātiti’a educational resources for students in an age range of 6-16 years old with a focus on coral reproduction or general topics surrounding coral reefs.
- We planned to have Terava come help with some measurements and physiological analyses in the lab this upcoming week between October 10 and 14th.
- Discussed possible future collaboration for helping you prepare documents for graduate school, help us write and develop our papers in the future for co-authorship, and work with us through some further data analysis.
- Discussed the stipend amount of $1,000 USD total for the amount of work you contribute, which can be split by the number of hours and amount that you prefer and you can bring this to us at the next meeting.
E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements of E5 tanks between 1230 and 1300. Conditions were cloudy so light levels in all tanks were low (<200 PAR). The paper clips are continuing to rust, so these will need to be replaced soon with permanent coated wire clips. Ariana is working on making these permanent clips. No mortality is observed. Daily measurements are recorded in GitHub here.
Airbrushing baseline fragments
All fragments sampled during the baseline sampling on 2 October 2022 were airbrushed today between 1630 and 1830. Ariana airbrushed with the following proceedure:
- Remove 4 frags at a time from the freezer and store them in a cooler on frozen beads. Label 50 mL falcon tubes for each fragment. Falcon tubes were labeled with “E5 Apul Metabolism; Oct 2022 Baseline; Airbrushed; Putnam; Frag ID”.
- Each fragment was airbrushed with chilled PBS buffer into a zip bag. The tissue slurry in the bag was then poured into the respective falcon tube. The homogenate volume was recorded in GitHub here and written on the falcon tube.
- The homogenate was placed on ice until all 4 frags were completed.
- After airbrushing, the homogenate was put in the -40C freezer.
- The process was repeated for all 24 fragments.
- After airbrushing, all equipment was cleaned with DI water and alcohol wipes.
- Airbrushed skeletons were placed in plastic cups with 10% bleach overnight to clean the skeletons. These will be kept at room temperature until wax dipping to determine surface area.
Notes: Note that before aliquoting for assays, the homogenate will need to be homogenized after thawing. Additionally, some of the fragments were quite small, so homogenate volume was low (4-7 mL). We will need to decide which physiology metrics we would like to measure first to make sure we have enough homogenate.
Logger calibration
Ariana wrote a script to read the light and temperature logger calibration files from 25 September 2022. The script can be found here on GitHub. This script reads in files in a loop and then conducts a linear model for each logger to the standard. For temperature, we used the standard as the average of all black Hobo loggers. These loggers are higher resolution than the Hobo pendants. By using the average value of these loggers we can calibrate any offset between loggers as well as calibrate the pendant loggers to a standard. For light, the Hobo pendant loggers (measuring light in Lux) are calibrated to the Odyssey light logger (measuring light in Integrated PAR) to allow us to convert Lux to Integrated PAR for analysis.
This script outputs files with the logger serial number, coefficient, and intercept values. These files can then be read in when analyzing light and temperature data to conduct calibrations in later analyses.
3 October 2022
Histological decalcification
Following the histological processing protocol Danielle taught Pierrick how to check the decalcification process for the September 2022 samples. Almost all of the samples besides 5 of them were ready to be resuspended in their original formalin tubes.
A. pulchra high frequency sampling dissections
Danielle finished processing the A. pulchra samples from the fourth high frequency sampling (20221002) for dissections. The fragments from this timepoint had many more distinct gonads.
E5 adult coral metabolism project
Tank cleaning
Ariana cleaned the E5 tanks today between 1100-1400. Tanks were cleaned by first removing the wire lines with frags attached from 1-2 tanks at a time and placing in the water table. The tanks were then removed and wiped with microfiber towels and rinsed with fresh water.
Fishing line of each fragment was cleaned and algae removed. Tanks were refilled and frags were added back in along with loggers and pumps. Ariana also cleaned the water lines.
During cleaning, tank position within each water table was rearranged. All tanks are in a new location. Frags were also returned such that each wire was put back in a different position to shuffle the fragments.
No mortality was observed in fragments. Water flow was adjusted to return to previous levels (350-400 mL per 5 sec).

Daily measurements
Daily measurements were taken between 1400 and 1500 and recorded in GitHub here. Conditions were cloudy and rainy today so light levels were low (<200 PAR).
Preparing for coral spawning - symbiont integration project
Symbiont density and innoculation trials
Ariana conducted symbiont isolation and counting trials to estimate how many fragments will be needed to innoculate and dose larvae with symbionts to allow for symbiont acquisition.
Ariana followed this procedure:
- Colelcted 3 test fragments from water table.
- Airbrushed coral fragment using PBS to remove tissue and pour into 50 mL falcon tube. Volumes were 10-15 mL of slurry after airbrushing.
- Homogenize - today this was done with pipetting and shaking the tube. In the full protocol use the microtip homogenizer for full homogenization.
- Spin to generate a symbiont pellet. Spun today in a 50 mL centrifuge (accuSpin 3R). For future protocols, convert or set in g force. Spun at 3500 rpm for 3 min.
- Pour out the supernatant and add 10 mL of PBS. Shake and homogenize. In innoculations, use filtered seawater rather than PBS.
- Spin again.
- Pour out supernatant and add 10-15 mL of PBS. Homogenize to resuspend symbionts. The symbiont slurry is now ready for counting cell density.
- Note total volume in each tube.
- Take 8 uL samples and load onto the hemocytometer. Today, four replicates were counted for each sample.
- Imaged hemocytometer at 10X and counted cells until reaching 100 cells. Only 1 square needed to be counted. The counts for each sample are shown below.
| Date | Time | Colony | Total Volume mL | Squares Counted | Count1 | Count2 | Count3 | Count4 | Mean Count | Cells per mL | Cells total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20221003 | 1600 | Test1 | 15 | 1 | 133 | 136 | 111 | 134 | 128.5 | 1285000 | 19275000 |
| 20221003 | 1600 | Test2 | 10 | 1 | 150 | 153 | 165 | 180 | 162 | 1620000 | 16200000 |
| 20221003 | 1600 | Test3 | 10 | 1 | 113 | 112 | 133 | 98 | 114 | 1140000 | 11400000 |
- Use C1 * V1 = C2 * V2 to solve for the volume of symbiont slurry that would be needed to dose symbionts at 3 x 10^4 as described in previous publications (e.g., Bay et al. 2011, Hartmann et al. 2018).
| Date | Tank | Symbiont Density (C1) | Target Density (C2) | Volume Symbionts (V1) | Tank volume (V2) | Time Added/Water Off | Time Water On | Notes |
|---|---|---|---|---|---|---|---|---|
| 20221003 | Test - frag 1 | 19275000 | 30000 | 28.0155642 | 18000 | NA | NA | Test density |
| 20221003 | Test - frag 2 | 16200000 | 30000 | 33.33333333 | 18000 | NA | NA | Test density |
| 20221003 | Test - frag 3 | 11400000 | 30000 | 47.36842105 | 18000 | NA | NA | Test density |
From these calculations, we will need more than 1 fragment if fragments are the same size as today per squarical. We have 12 squaricals to innoculate. In the experiment, we will therefore use larger fragments and use 1-1.5 fragments per squarical for innoculation at the target density. The innoculation period will be 3 days with once daily innoculations, so we will need 36-52 total fragments from our parent population of approx. 20-30 colonies.
2 October 2022
A. pulchra high frequency sampling - 20221002
Completed the fourth timepoint for the high frequency project for A. pulchra on the samples below. Went in the field at 09:00 to collect 20 fragments from the five patches.
Metadata:
| Species | Site | OrderbyPatch | Original.Tag.Num | Patch | Latitude.S | Longitude.W | GPS.Point.Name | Timestamp | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. pulchra | Mahana | P1.3 | 414 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:59 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.5 | 416 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:11 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.4 | 424 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:17 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.1 | 426 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:49 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.1 | 429 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:05 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.4 | 430 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:10 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.9 | 431 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:46 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.5 | 433 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:02 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.4 | 434 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.10 | 436 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:29 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.2 | 437 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:50 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.10 | 439 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:47 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.1 | 448 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:35 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.2 | 449 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:07 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.8 | 451 | P5 | 17°29.220’ | 149°53.236’ | P5 | 2:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.6 | 458 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.11 | 461 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:30 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.1 | 462 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.6 | 464 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:41 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.6 | 465 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:22 PM |
A. pulchra high frequency sampling protocol
Danielle followed the same day sample processing protocol for the 20 collected high frequency samples. Danielle also set aside 10 fragments to be dissected and preserved in DNA/RNA shield.
Dissections
Danielle started processing the A. pulchra samples from the fourth high frequency sampling (20221002) for dissections. The fragments from this timepoint had many more distinct gonads.
E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements at 1200-1230 in all E5 tanks. Recorded in GitHub here. Flow was even across tanks (350-375 mL per 5 sec) in each tank and did not need to be adjusted. Tomorrow the team will clean and rearrange all tanks according to the weekly maintainance protocol.
Baseline adult sampling
Ariana sampled adult fragments for baseline measurements as described in the baseline measurement preparation post on 30 September 2022. Sampling followed the baseline measurement protocol.
First, the selected fragments (N=24, n=4 per genotype) were removed from the tanks and imaged using a color standard and ruler for reference.

Fragments were then transported to the molecular lab for processing. As described in the protocol, each fragment was sampled for biopsies that were flash frozen (n=2 per frag) and stored in RNA DNA shield (n=2 per frag) with the remaining fragment frozen for physiology. Between samples, equipment was sterilized with 10% bleach followed by 70% ethanol. Gloves and counter were sterilized between each fragment. Sterilization was critical since these samples may be used for bacterial community characterization.
For each fragment, pre-labeled tubes and bags were used as described on 30 September 2022 post. Using bone cutters, 4 biopsies (scraping 5-10 polyp area) were taken and added to tubes. Flash frozen tubes were immediately frozen in liquid nitrogen. Tubes with RNA DNA shield were stored in the -40C freezer. The remaining fragment was stored at -40C. After all samplinig was completed, all tubes were then stored in the Gump molecular lab freezer at 40C.
Sampling of all fragments took 1.5-2 hours. Sampling of fragments was conducted in random order to avoid any time effects on genotype differences.
Checklists for sampling and sample inventory are available on GitHub.
Downloading loggers
HOBO Pendant loggers from the six E5 tanks were downloaded at 1220. Loggers were kept logging in the tanks. Black HOBO loggers from the wildtype parent tanks (n=2) were removed from tanks, stopped, and downloaded at 1030. Logger files can be found here.
Histological decalcification
Following the histological processing protocol Danielle taught Pierrick how to preserve and decalcify his Pocillopora spp. samples and she began the decalcification process for the September 2022 samples. They were put into the pre-labeled March and June cups.
A. pulchra high frequency sampling - 20220927
Completed the third timepoint for the high frequency project for A. pulchra on the samples below. Went in the field at 09:00 to collect 20 fragments from the five patches.
Metadata:
| Species | Site | OrderbyPatch | Original.Tag.Num | Patch | Latitude.S | Longitude.W | GPS.Point.Name | Timestamp | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. pulchra | Mahana | P1.3 | 414 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:59 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.5 | 416 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:11 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.4 | 424 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:17 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.1 | 426 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:49 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.1 | 429 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:05 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.4 | 430 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:10 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.9 | 431 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:46 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.5 | 433 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:02 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.4 | 434 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.10 | 436 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:29 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.2 | 437 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:50 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.10 | 439 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:47 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.1 | 448 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:35 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.2 | 449 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:07 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.8 | 451 | P5 | 17°29.220’ | 149°53.236’ | P5 | 2:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.6 | 458 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.11 | 461 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:30 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.1 | 462 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.6 | 464 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:41 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.6 | 465 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:22 PM |
A. pulchra high frequency sampling protocol
Danielle followed the same day sample processing protocol for the 20 collected high frequency samples. Danielle also set aside 10 fragments to be dissected and preserved in DNA/RNA shield.
Dissections
Danielle continued processing the A. pulchra samples from the second high frequency sampling for dissections. Danielle and Pierrick began to work on the Pocillopora spp. dissections and found where the eggs/gonads should be in the Pocillopora spp. tissue (right below the polyps). Since they are so small (~60um) they began the fixing and decalcification process for a subset of samples to see if any reproductive tissue can be seen in a scope after decalcification.
Tissue biomass completion - September 2022 gametogenesis project
Danielle taught Pierrick the final step in the tissue biomass protocol and weighed the ash pans of the host and symbiont portions of each of the September 2022 gametogenesis timeseries samples (n=40) with 4 mL for host and 1 mL for symbionts in the pans.
1 October 2022
The team took the day off today for some fun snorkeling at Temae!
E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements of all tanks at 1630-1645. Sunlight was limited today and it was cloudy/rainy. Light measurements were all low during measurements (<25 PAR). Recorded in GitHub here. Flow was even across tanks (325-375 mL per 5 sec) in each tank and did not need to be adjusted. Ariana will check the outflows tomorrow and flush the lines to prevent lines clogging after sampling the adult baseline fragments.
30 September 2022
Gamete dissection
Danielle continued her methods for dissecting gametes from live A. pulchra fragments.
Danielle used a bone saw to cut ~10 individual <1 cm pieces from each individual fragment and submerged them into a microscope dish under a microscope light to identify if any gonads were present. She then used fine tipped tweezers to hold the fragment place and used a 1mL pipette to lightly dislodge the gonads. She then used a glass pipette to apply pressure and suction to the gonad until it finally released from the tissue. She then preserved the gonads she was able to collect from 5 separate fragments into 250 uL of DNA/RNA shield in a sterile o-ring micro-centrifuge tube. She was also able to identify gonads from other parts of tissue as they float when dislodged.
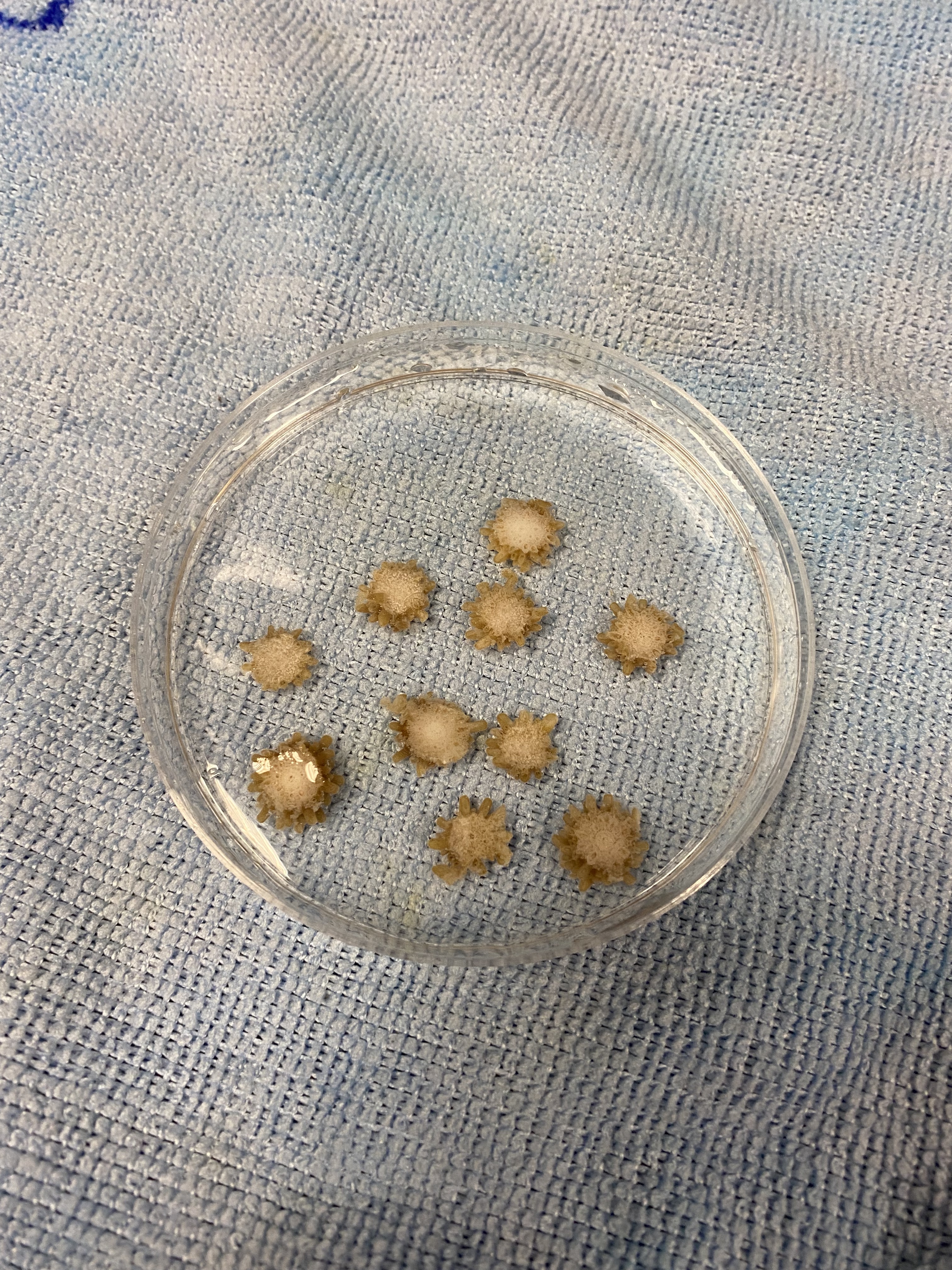


E5 adult coral metabolism project
Daily measurements
Ariana took daily measurements at 0945-1020. Fragments look clean and healthy, no mortality observed. Flow adjusted to be between 350 and 400 mL per 5 sec in all tanks.
Daily measurements for today were recorded in GitHub here.
Preparing adult sampling
In the next couple of days Ariana will sample n=4 randomly selected fragments of each genotype to characterize baseline values of our responses of interest including DNA methylation and chromatin structure, gene expression, metabolomics, microbial communities, and physiology. Fragments were selected by randomly selecting n=4 per genotype such that each fragment was from a different tank and sampling from each tank was balanced to keep fragment density consistent.
Today, Ariana identified the fragments to be sampled and prepared and labeled all tubes, whirlpacks, and equipment that will be used.
Baseline sampling will be conducted following the protocol written by Ariana today. The baseline sampling protocol details procedures taking photographs of selected fragments for gene expression, microbial communities (16S and ITS2), chromatin structure and DNA methylation, metabolomics, and physiology.
The fragments that will be sampled are:
A7, 16, 20, 31
B9, 20, 25, 34
C1, 19, 26, 30
D4, 13, 26, 30
E5, 8, 20, 31
F2, 9, 17, 22
Labeling equipment
2 mL screw cap cryo tubes were labeled in preparation for sampling as nubmers 1-96. The number was written on the top cap of each tube as well as on the side. “E5 Metab. 2022” was also added on the side of each tube to identify the project. To see details on organization and labeling for baseline sampling, see the detailed protocol here.
Tubes #1-48 were filled with 1 mL of RNA DNA Shield. All tube ID’s and associated fragment and method of preservation were planned and recorded in the sample inventory.
Ariana also created a fragment tracking datasheet and baseline sampling checklist that will be used to track sampling and metadata for this project.
Two tube boxes were labeled as well with one box for RNA DNA shield samples (Tubes 1-48) and snap frozen samples (Tubes 49-96).
Finally, n=24 whirlpacks were labeled with fragment number as well as project information (“E5 Apul metabolism; Oct 2022 baseline; Putnam”).
Documents referenced in this post are all found below:
29 September 2022
Chlorophyll - September 2022 gametogenesis project
Danielle taught Pierrick the final steps in the chlorophyll protocol and completed the 40 September 2022 gametogenesis timeseries samples chlorophyll analysis.
Tissue biomass preparation - September 2022 gametogenesis project
Danielle taught Pierrick the second step in the tissue biomass protocol and weighed the pre-burned pans and filled the host and symbiont portions of each of the September 2022 gametogenesis timeseries samples (n=40) with 4 mL for host and 1 mL for symbionts in the pans. They put the aluminum pans in the drying overn for >24 hours.
E5 adult coral metabolism project
Ariana took daily measurements of the adult corals. The protocol for daily measurements can be found here.
The daily probe set up looks like the photo below. We take measurements of water flow, temperature (C), pH (mV), conductivity (psu), and light (PAR). Daily measurements except for light (see below) are taken at the center of each tank simultaneously.

Light is taken in the following locations in each tank:
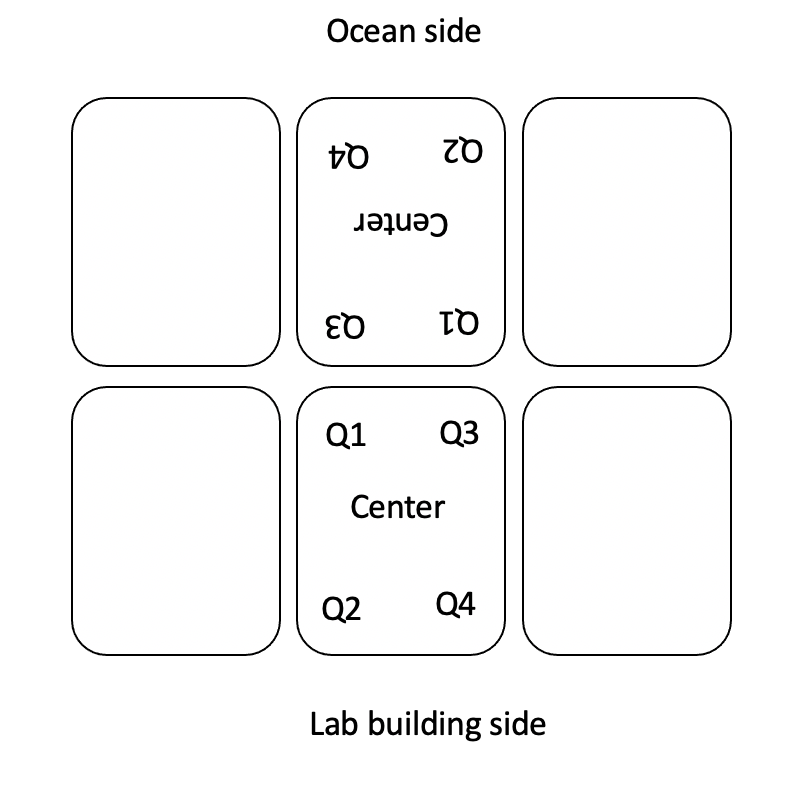
Daily measurements for today were recorded in GitHub here.
All tanks and fragments looked good today and no mortality was observed. Ariana is going to start making materials to replace the paperclips with coated wire. After about 1 week they will be rusting.
Physiology processing
Danielle processed physiological samples for ash free dry weight and chlorophyll content.
28 September 2022
Chlorophyll preparation - September 2022 gametogenesis project
Danielle taught Pierrick the first step in the chlorophyll protocol and prepped the 40 September 2022 gametogenesis timeseries samples with acetone. All samples put in dark fridge in molecular at 09:05.
Tissue biomass preparation - September 2022 gametogenesis project
Danielle taught Pierrick the first step in the tissue biomass protocol and labeled and prepped 80 aluminum pans labeled 1-80 for the two host and symbiont portions of each of the September 2022 gametogenesis timeseries samples (n=40). She also put the aluminum pans in the muffle furnace for the pre-burning step in the protocol.
Pocillopora spawning
Ariana and Pierrick snorkeled to observe potential Pocillopora spp. coral spawning from 0530-0800 at Manava site. No spawning was observed.
Cleaning tanks
The team cleaned all large blue tanks today. Due to a clog in the line, water flow was cut off to one blue tank earlier this week resulting in some coral mortality. Dead portions of colonies were cut off of colonies and removed from tanks. All tanks were scrubbed and cleaned and filled with new water. Colonies will be taken back to the reef on Friday now that E5 fragmentation is complete (see below).
E5 adult coral metabolism experiment
Ariana cleaned all tanks, cleaned all fragments, secured clips to wire hangers, and finished labeling and fragmenting corals. Danielle assisted with fragmentation and labeling. These are described in more detail below.
Ariana has also started documents for entering data while tracking daily environmental measurements, conducting daily measurements and weekly cleanings, and tracking fragment survival in the GitHub repository. Ariana will write and post links to protocols here.
Fragmentation
Ariana finished fragmenting corals today for this project. There was high mortality of fragments for genotype C that was fragmented on 26 September 2022 (8 fragments showed mortality). Therefore, all C fragments were removed and replaced with a new genotype. This was also done for genotype A in which >80% of fragments showed signs of mortality. There was no other observed mortality in fragments from genotypes B, D, and E.
New genotypes for A and C fragments and for F fragments were fragmented at 1600-1800. 35 fragments were obtained from each genotype and were fragmented as previously described below. Fragments were attached to labels and added to tanks by 2100. The fragments varied in size from 2 cm to 6 cm in length depending on the available fragments from the morphology of the parent colony.
The final list of tank fragment locations is available on GitHub and listed below.
Tank 1:
- A1-A5
- B1-B5
- C1-C6
- D1-D6
- E1-E6
- F1-F6
Tank 2:
- A6-A11
- B6-B11
- C7-C11
- D7-D11
- E7-E12
- F7-F12
Tank 3:
- A12-A17
- B12-B17
- C12-C17
- D12-D17
- E13-E17
- F13-F18
Tank 4:
- A18-A23
- B18-B23
- C18-C23
- D18-D23
- E18-E23
- F19-F24
Tank 5:
- A24-A29
- B24-B29
- C24-C29
- D24-D29
- E24-E29
- F24-F29
Tank 6:
- A30-A35
- B30-B35
- C30-C35
- D30-D35
- E30-E35
- F30-F35
In the case that another genotype experiences mortality, there are 2 extra genotypes that were fragmented and will be held in the water tables.

Cleaning and tank set up
Ariana cleaned all treatment tanks by removing fragments and placing them in the water table with running water. Tanks were wiped clean and rinsed and re filled. Strings on fragments were cleaned to remove algae.
Ariana made notches in the hanging wires to prevent clips from moving around and the corals from getting too close to each other. These have worked well today.
In the next week, we will switch the paper clips to plastic clips to prevent rusting.


Feeding treatments
In 27 September 2022 post, Ariana described potential feeding treatments using artificial food. Ariana measured tank dimensions today for the E5 project - 40 cm W x 55 cm L x 30 cm H = ~66 liters. This is equivalent to about 18 gallons.
Reef Chili: Manufacturer recommends 1 scoop per 20 gallons. We would use 1 scoop per feeding per tank = 4 scoops per feeding total. If we assume 1 scoop = 1 gram, then we would have approx. 40 feedings per jar of reef chili (40 g per jar). If we feed 3x per week from mid-October to mid-December we will have 24 feedings. This will require One 1.4oz container has approximately 100 scoops. We should order 5 jars of reef chili to be safe. It can be stored in the fridge for up to a year if we do not use it. It looks like this item is currently out of stock.
Reef Roids: Manufacturer recommends 1 teaspoon per 100 gallons. We would therefore use 0.2 teaspoons per tank x 4 tanks = 0.8 teaspoons per feeding. With a total of 24 feedings, we would need ~20 teaspoons of Reef Roids. The bags are sold in units of 75 g or 150 g. On the website, it says “On average, the 30-gram bag of Reef-Roids will last roughly three months in a typical 50-gallon reef tank with bi-weekly feeding.”. If we calculate this out, bi weekly feeding of a 50 gallon tank for 3 months would use 0.5 teaspoons per feeding x 2 feedings x 12 weeks = 12 teaspoons in a 30 gram bag. If we need 20 teaspoons minimum, we would need to order the 75 g bag. To have extra, we should order the 150 g size. It can be refrigerated to last longer if we do not use it.
27 September 2022
Pocillopora spawning
Danielle and Pierrick snorkeled to observe Pocillopora spp. coral spawning at Manava from 0530-0730. No spawning was observed.
A. pulchra high frequency sampling - 20220927
Completed the second timepoint for the high frequency project for A. pulchra on the samples below.
Metadata:
| Species | Site | OrderbyPatch | Original.Tag.Num | Patch | Latitude.S | Longitude.W | GPS.Point.Name | Timestamp | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. pulchra | Mahana | P1.3 | 414 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:59 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.5 | 416 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:11 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.4 | 424 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:17 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.1 | 426 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:49 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.1 | 429 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:05 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.4 | 430 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:10 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.9 | 431 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:46 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.5 | 433 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:02 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.4 | 434 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.10 | 436 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:29 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.2 | 437 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:50 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.10 | 439 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:47 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.1 | 448 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:35 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.2 | 449 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:07 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.8 | 451 | P5 | 17°29.220’ | 149°53.236’ | P5 | 2:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.6 | 458 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.11 | 461 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:30 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.1 | 462 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.6 | 464 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:41 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.6 | 465 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:22 PM |
A. pulchra high frequency sampling protocol
Danielle and Pierrick followed the same day sample processing protocol for the 20 collected high frequency samples. Danielle also set aside 10 fragments to be dissected and preserved in DNA/RNA shield.
Boating
Ariana was officially checked out on Gump LTER boats. Ariana can now drive the boats to our field sites.
E5 adult coral metabolism experiment fragmentation
Ariana fragmented two more genotypes for use in the E5 adult experiment as described on 26 September 2022. Genotypes fragmented today were labeled D1-D35 and E1-E35. Fragments from colony A were showing mortality. These were removed from the tanks and will be replaced with a different genotype tomorrow. Fragmentation was completed from 0700-0900. Ariana and Danielle created labels in preparation for the final fragmentation tomorrow.
Fragments from yesterday appear to be healthy and the tank system is working well. In about a week we will need to change the paperclips to plastic coated paperclips to avoid rusting.
Once fragmentation is completed, Ariana will start daily measurements of environmental conditions, regular tank cleaning, and final treatment preparations.
The experimental design has been finalized for this project as is as follows:
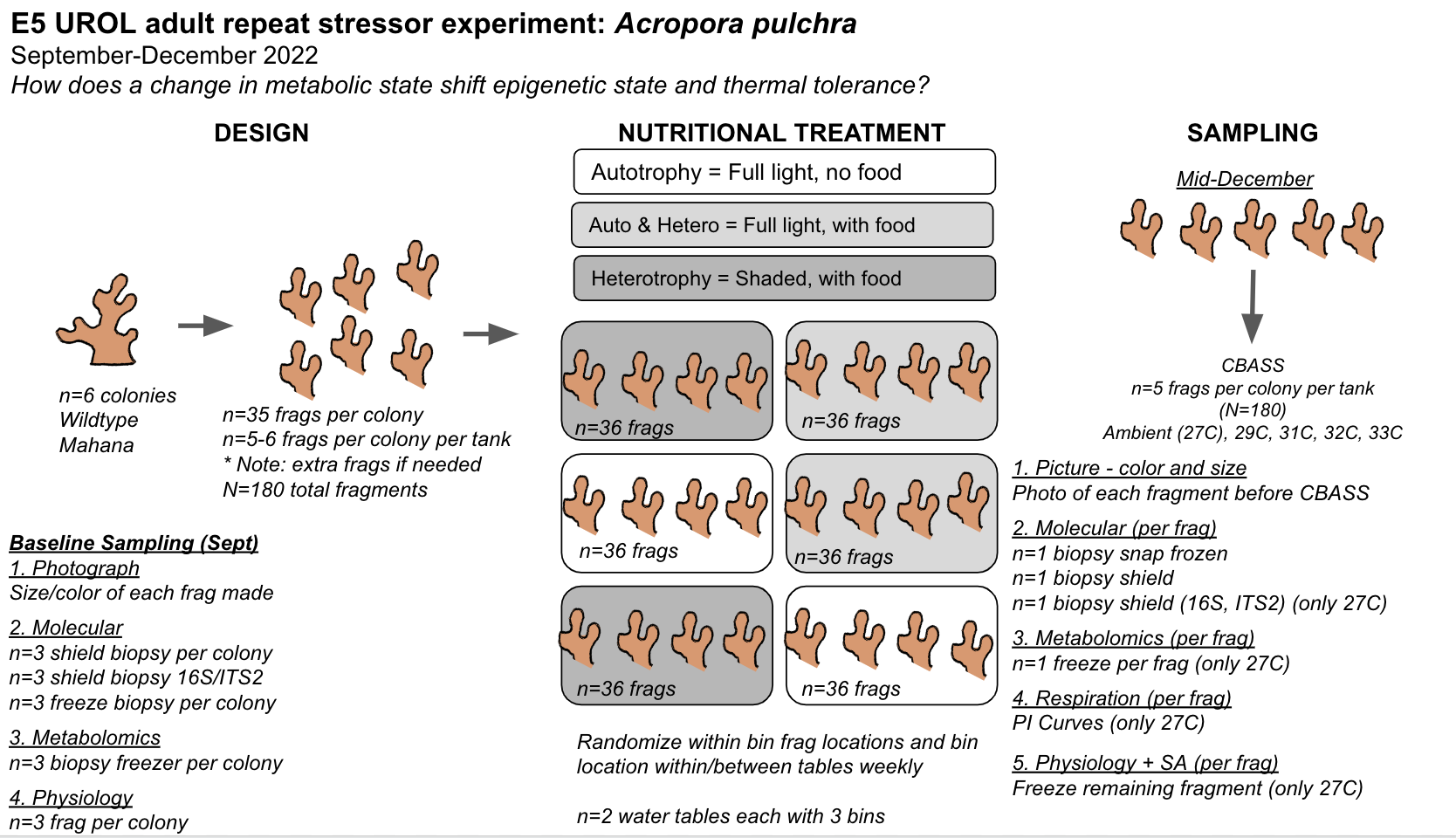
This updated design includes a final processing for all fragments that includes the following responses:
- All fragments (5 per colony/genotype per tank) undergo CBASS testing at 5 temperatures including 27C, 29C, 31C, 32C, 33C (or something similar).
- We will take molecular samples from all fragments after CBASS.
- The fragment at the control temperature will then be used for molecular, metabolomic, and physiology responses.
Feeding treatments
For this experimental design, we need to plan for the type and amount of food to feed for Hollie to bring when she arrives in Moorea. Ariana looked in the literature for information on the effect of artificial foods (freeze dried plankton, rotifers, etc.) on coral growth.
Conlan et al. 2018: This paper compared coral growth with different types of food in A. millepora and P. acuta. They found no significant effect of feeding artificial foods on growth when compared to raw seawater. The dosage used was 0.05 g per tank in 49 L tanks. This equals about 1 mg per litre water.
Forsman et al. 2012: This paper examined the effects of different aritifical foods and dosage of foods on growth in M. capitata, P. acuta, and P. compressa. They tested 1X, 3X, and 10X of manufacturer’s dosage of Reef Roids, Reef Chili, and several other brands of food. Specifically, in M. capitata (in the Acroporidae family) has positive growth responses to artificial foods. They found that dosage of 1X of manufacturer’s instructions of Reef Roids or Reef Chili were best for this species. It is important not to overfeed, as this can reduce growth and cause tank algal overgrowth.
For feeding in this experiment, Ariana suggests that we purchase either Reef Roids or Reef Chili. We can feed corals 3X per week in treatment tanks. Water can be turned off for 1 h during feeding. Ariana will look into the amount of food that will be required for this feeding. If the amount is too high, we can look into building a feeding tank for all corals to be fed as a group and thereby reduce the amount of food needed.
Remaining to-do items for experiment set up
- Prepare a tank for 3x week feedings
- Remake paper clips to plastic clips
- Create a schedule document for daily measurements and weekly cleaning check offs
- Start treatment after ~ 2-week recovery period
- Order coral food
26 September 2022
Logistics - boating and liquid nitrogen
Danielle reserved boat days for the blue boat for returning and collecting colonies for spawning and after spawning. She also arranged for the next round of liquid nitrogen to be ordered and returned by Friday, the 29th.
Aliquoting gametogenesis project - September 2022
Danielle taught Pierrick the aliquoting protocol for Mo’orea and also aliquoted 40 of the September 2022 monthly sampling A. pulchra fragments following the protocol and stored the remaining tissue homogenate and samples in the Molecular -40 freezer in a mesh bag.
Surface area gametogenesis project - September 2022
Danielle taught Pierrick the coral surface area protocol for Mo’orea and also completed the standard curve for surface area and measured surface area 40 of the September 2022 monthly sampling A. pulchra fragments following the protocol and stored the wax dipped skeletons in the molecular lab in a zip lock bag.
Pocillopora spawning
Ariana and Pierrick snorkeled to the Manava field site at 5:30-7:00am to observe potential Pocillopora spp. spawning. No spawning was observed. This site has 100m squared area with POC tags that Pierrick is observing and sampling for his project. Tomorrow we will stay in the water longer. We will observe for spawning in Oct - Dec on nights 0, 1, and 2 after the full and new moons for 45min-2h after sunrise.
Logger deployment
Ariana deployed pendant loggers for the E5 adult project. The following loggers were deployed. Tanks were numbered as 1-6 for simplicity.
| SSN | Interval | Time in Tanks | Tank |
|---|---|---|---|
| 21335989 | 10 min | 1115 | Tank 1 |
| 21002980 | 10 min | 1115 | Tank 2 |
| 21002972 | 10 min | 1115 | Tank 3 |
| 21335984 | 10 min | 1115 | Tank 4 |
| 20937873 | 10 min | 1115 | Tank 5 |
| 21002976 | 10 min | 1115 | Tank 6 |
| 20444033 | 10 min | 1115 | Wild parent tank 1 |
| 20719656 | 10 min | 1115 | Wild parent tank 2 |
Fragmentation
Today the team started the process of fragmenting adult A. pulchra corals for the E5 project to investigate the effects of altered metabolic state on epigenetic state and thermal performance.
To fragment, the team made and labeled strings (fishing line) attached to paper clips with lab tape as a label on the paper clip. These were then tied to coral fragments on the upper part of the branch such that the branch was oriented up right. The fragments were then clipped onto the wire using the paper clips. Fragments were suspended 15-18 cm below the water surface.
Coral fragments (~4-8 cm) were cut using bone clippers from larger colonies. Fragmentation was done in a water table from 1100-1900.

All treatment tanks were cleaned and pumps were added into each tank. Hobo pendant loggers are on the bottom center of each tank. Flow is high for these tanks to help corals recover from fragmentation. Note that no treatments have started yet, after fragmentation there will be a recovery period.


Fragments were then allocated to tanks with tank number recorded in GitHub here. Each tank has 5-6 fragments per colony and will have equal numbers of fragments.
A total of 3 genotypes/colonies were fragmented today and the team pre-labelled strings and paperclips for tomorrow. Ariana will finish fragmenting tomorrow and baseline sampling wll take place on Thurs/Friday of this week.
Fragmented today:
Colony A: Fragments A1-A35
Colony B: Fragments B1-B35
Colony C: Fragments C1-C25
Note that A fragments have some mortality. Ariana may switch out colony A for a different colony tomorrow. Water quality and flow was poor in this tank today that may have led to some of the mortality.
Tomorrow we will fragment colonies D, E, F.
Tank assignments
The tanks for each fragment are listed below and recorded in GitHub (see link above).
Tank 1:
- A1-5
- B1-5
- C1-6
Tank 2:
- A6-11
- B6-11
- C7-11
Tank 3:
- A12-17
- B12-17
- C12-17
Tank 4:
- A18-23
- B18-23
- C18-23
Tank 5:
- A24-29
- B24-29
- C24-29
Tank 6:
- A30-35
- B30-35
- C30-35
Revised E5 design
Given the amount of space in each tank and the number of fragments we could get per colony, Ariana re calculated our E5 design numbers. The changes are:
- Removing mid November time point. Since we want to know about how metabolic state influences stress response, it makes the most sense to keep our replication/samples for the time point in which we plan to do CBASS.
- Changed treatments to include autotrophy (light, no food), auto/hetero (light, added food), and heterotrophy (shaded, no food) as suggested by R. Cunning. This will provide nutritional treatments to shift metabolic state. Given our current design, we can do n=2 tanks per treatment.
- tanks will now include 5-6 fragments per colony. This will provide enough samples to take baseline samples this week as well as have 1 fragment for physiology and molecular and 3-4 fragments for CBASS. We can destructively sample following CBASS. The temperatures for CBASS will be chosen to represent a control, moderate heating, and high heating tempratures.
- Colony number has been reduced from 10 down to 6. There is not enough room in the tanks for all 10 colonies.
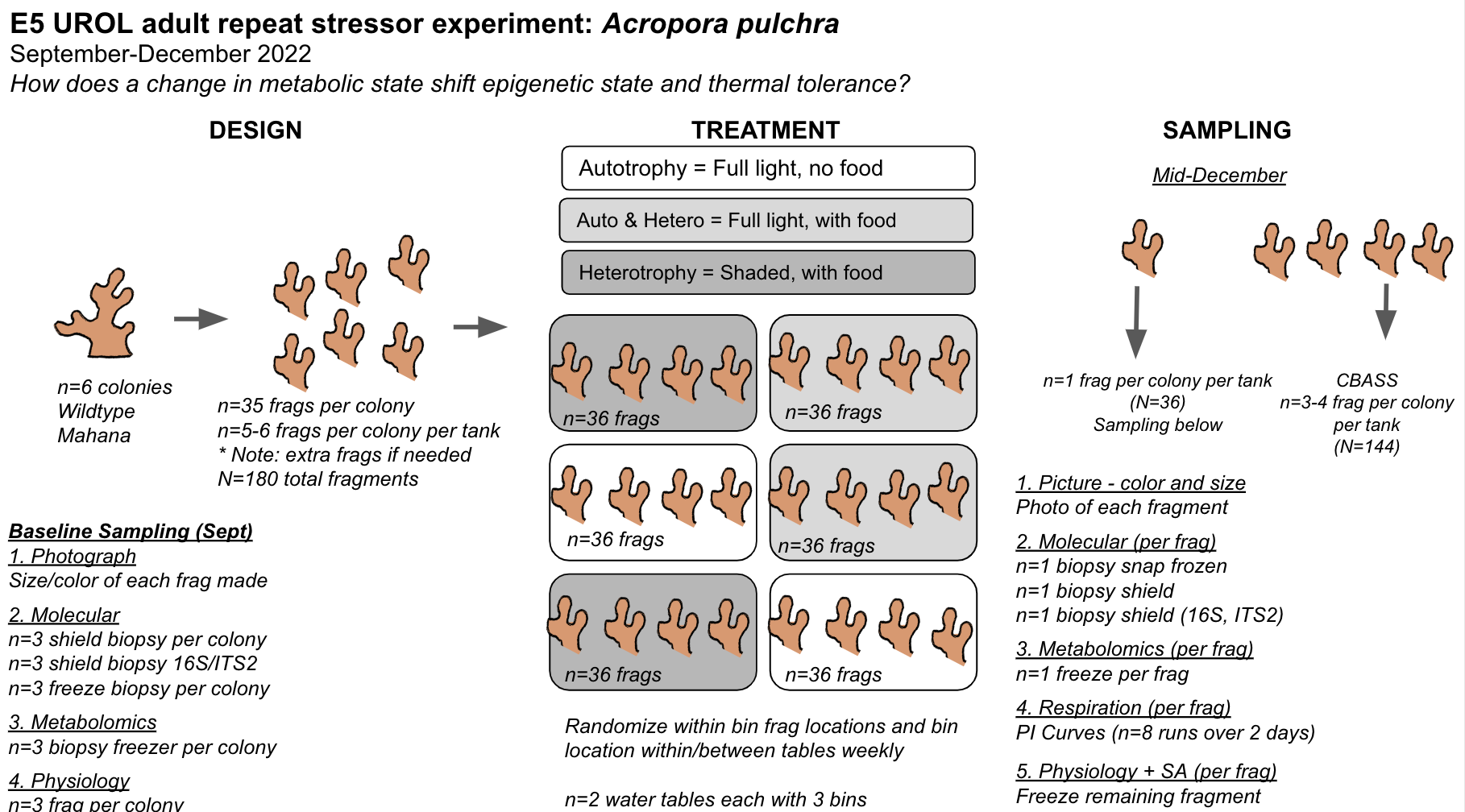
Boating
Over the last few days, Ariana has completed the required boating trips and CA online boating course. Ariana will work with Katherine to check out on the Gump boats (509).
25 September 2022
Logger calibration
Ariana removed loggers from tanks at 1030 and downloaded all logger files at 1200 and began writing a script to calulate the offsets for each logger.
Tank set up
Ariana set up the E5 tanks to have hanging wires for each fragment when fragmentation occurs tomorrow.


Planning for tomorrow
Tomorrow Ariana will fragment the A. pulchra corals. These are the steps we will follow to fragment:
- Select corals with the most fragments/branches (n=10 preferred).
- Place the first colony in a holding tank on the water table.
- Use bone cutters and a frag saw to cut as many fragments as possible approx. 3-4 in long at the branches.
- Label the fragments by attaching to a paperclip with a tape label using fishing line. Label with a colony letter and number (e.g., A1-A30, B1-B30, etc.).
- Place the fragments in the tanks randomly distrubted between the tanks.
- Return the remaining colony to the holding tank.
This fragmentation will be followed by baseline sampling at the start of the treatment period depending on the number of fragments available.
24 September 2022
Airbrushing gametogenesis project - September 2022
Danielle taught Pierrick the airbrushing protocol for Mo’orea and also airbrushed 40 of the September 2022 monthly sampling A. pulchra fragments following the protocol and stored the tissue homogenate in the Molecular -40 freezer in a mesh bag.
Logger calibration
Ariana added the two pendant loggers from the incubators into the calibration tank that were missed yesterday.
Logger 21335989 - logging every 10 min
Logger 21002976 - logging every 10 min
Added to calibration tank at 0900. Note that data from 0700-0830 may need to be removed from the calibration. At this time the morning sun is causing shadows in the tank that may make the calibration in accurate.
Daily measurement probe set up
Daily measurements for larval and the E5 adult project should include:
- Flow rate (mL / 10 sec): Use a graduated clyinder to measure the amount of water from the input tube in a 10 sec period.
- Temperature (C): Measurement of temperature in the center of each tank.
- Light (PAR): Measurement of light at the center of the tank and in the center of each of 4 quadrants (total of 5 measurements per tank).
- pH and salinity: Measure pH and salinity with the Orion probe at the center of each tank.
Time of day, tank number, and notes regarding weather or other conditions should be noted.
For Ariana’s larval project, daily measurements will be recorded in Google Drive here.
For the E5 adult project, daily pH, temperature, and flow measurements will be recorded in GitHub here and daily light measurements will be recorded in GitHub here.
The protocol for taking daily measurements was written by Ariana available here in the E5 protocols repository based on the Putnam Lab protocol.
Ariana prepared all probes and measured a standard curve for the pH probe across a range of temperature with Tris using the Putnam Lab protocol. 15 mL of Tris was added to a falcon tube and placed in the fridge for ~20 min. The temperature and pH probe were then submerged into the falcon tube on the bench top. The calibration started at 11°C and slowly warmed to room temperature with measurements of temperature and Tris every ~1°C. Once the Tris reached room temperature the Tris and probes were moved into a benchtop incubator (MyTemp Mini) and warmed to the upper temperature used in the calibration (~35C).
This data is below and was recorded in Ariana’s Google Drive here and in the E5 repository here.
| Date | Time | Temp.Tris | mV.Tris |
|---|---|---|---|
| 20220924 | 1013 | 11.18 | -73 |
| 20220924 | 1016 | 13.31 | -72 |
| 20220924 | 1020 | 14.66 | -70.3 |
| 20220924 | 1025 | 16.18 | -68.3 |
| 20220924 | 1030 | 17.28 | -66.7 |
| 20220924 | 1049 | 18.75 | -64.9 |
| 20220924 | 1057 | 19.48 | -64.1 |
| 20220924 | 1102 | 20.18 | -63.1 |
| 20220924 | 1106 | 21.08 | -61.9 |
| 20220924 | 1111 | 22.19 | -60.6 |
| 20220924 | 1115 | 23.07 | -59.4 |
| 20220924 | 1120 | 24.04 | -58 |
| 20220924 | 1125 | 25.03 | -56.7 |
| 20220924 | 1130 | 26.06 | -55.4 |
| 20220924 | 1135 | 27.27 | -53.9 |
| 20220924 | 1140 | 28 | -52.8 |
| 20220924 | 1145 | 28.96 | -51.8 |
| 20220924 | 1150 | 30.42 | -49.6 |
| 20220924 | 1153 | 31.06 | -48.6 |
| 20220924 | 1159 | 32.27 | -47.1 |
| 20220924 | 1204 | 33.35 | -45.6 |
| 20220924 | 1210 | 34 | -44.8 |
| 20220924 | 1215 | 35.12 | -43.1 |
| 20220924 | 1220 | 36.22 | -41.6 |
23 September 2022
Heatwave colony return
The team returned the heatwave A. pulchra colonies from Danielle’s project to the Mahana site from 0900-1100. We will collect them again before the next spawning. Colonies were zip tied to cinder blocks on Patch 3 at Mahana.

Adult E5 experiment set up
Ariana cleaned water lines and started water in n=6 tanks in preparation for the adult experiment.
Ariana started a GitHub repo for this project here.
Logger offload and calibration
Loggers were read off for the wildtype parent tanks and Danielle’s parent tanks. Files were labeled with SSN and date and saved in the Apulchra_metabolism E5 GitHub repository. Loggers read off were:
20719656 (Wildtype Tank 2)
20946644 (Danielle Tank 1)
20444033 (Wildtype Tank 1)
20444040 (Danielle Tank 2)
Ariana started a calibration of all temperature and light loggers that will run over the next 24-48 h. This will allow us to convert Lux to PAR from the Hobo Pendant loggers and to calculate temperature offsets from the temperature logging on the Hobo Pendants.
The following loggers were launched:
| Logger | Interval | Type | Time Deployed |
|---|---|---|---|
| 20719656 | 10 min | Hobo Temp | 1635 |
| 20946644 | 10 min | Hobo Temp | 1635 |
| 20444033 | 10 min | Hobo Temp | 1635 |
| 20444040 | 10 min | Hobo Temp | 1635 |
| 21335986 | 10 min | Pendant | 1635 |
| 21002981 | 10 min | Pendant | 1635 |
| 21002972 | 10 min | Pendant | 1635 |
| 20937872 | 10 min | Pendant | 1635 |
| 21002975 | 10 min | Pendant | 1635 |
| 21335980 | 10 min | Pendant | 1635 |
| 21335987 | 10 min | Pendant | 1635 |
| 21002980 | 10 min | Pendant | 1635 |
| 21002977 | 10 min | Pendant | 1635 |
| 21002974 | 10 min | Pendant | 1635 |
| 21335982 | 10 min | Pendant | 1635 |
| 21002982 | 10 min | Pendant | 1635 |
| 20937873 | 10 min | Pendant | 1635 |
| 21335983 | 10 min | Pendant | 1635 |
| 21335984 | 10 min | Pendant | 1635 |
| 21002979 | 10 min | Pendant | 1635 |
| 15643 | 10 min | Odyssey Light | 1635 |
The Odyssey logger was previously calibrated by Danielle Becker-Polinski with this protocol.
Loggers were arranged as pictured below in one tank with running water at a low flow to avoid surface disturbance. These loggers will run for 24-48 h to conduct the calibration for light and temperature data.
22 September 2022
Histology time series
We completed a sampling time point for the A. pulchra histology time series at the Mahana site (see details beow from 16 September 2022) for the high frequencing sampling. The purpose of this sampling is to acquire higher resolution histology from this species through the last month until spawning. This will occur every 5 days.
We sampled n=20 colonies (n=4 from each patch). We sampled between 0930 and 1100. We returned to the lab and processed the samples as described on 17 September 2022 between 1100 and 1230.
E5 project set up
Ariana continued setting up materials for the E5 adult stressor project. We purchased 6 bins that will be tanks for this experiment. Ariana prepared the bins by drilling drain holes and securing wire at the top of each tank to hang fragments.
Tetiaroa spawning preparations
The team met with collaborators in the Padilla-Gamino lab from the University of Washington to assist with preparations for a collaborative spawning expedition that will occur in October. The Padilla-Gamino lab will conduct spawning in Tetiaroa in October with parallel measurements in Moorea to characterize reproduction in Acropora corals. We shared protocols with their lab for spawning and measuring response variables of interest.
Gamete dissection
Danielle trialed methods for dissecting gametes from live A. pulchra fragments. During our field expedition today, we noticed that gametes were visible in A. pulchra fragments and Danielle dissected and sampled these gametes.
Danielle first cut extra fragments that were taken from sampling with bone cutters into a ~0.5 inch piece and submerged in a microscope dish under a microscope light to identify if any gonads were present in the fragment piece. She then used fine tipped tweezers to hold the fragment piece and used a fine pointed dissecting tool to gently dislodge any of the gonads from the underlying tissues. She then used a glass pipette to apply pressure and suction to the gonad until it finally released from the tissue. She then preserved the 9 gonads she was able to collect from 5 separate fragments into 250 uL of DNA/RNA shield in a sterile o-ring micro-centrifuge tube. This process will be modified next sampling period to try a fragment saw and needle syringe to improve the number of gonads extracted from samples. The below images show clear gonads (small white circular material in the skeleton)located in the polyp pockets of the fragment pieces.
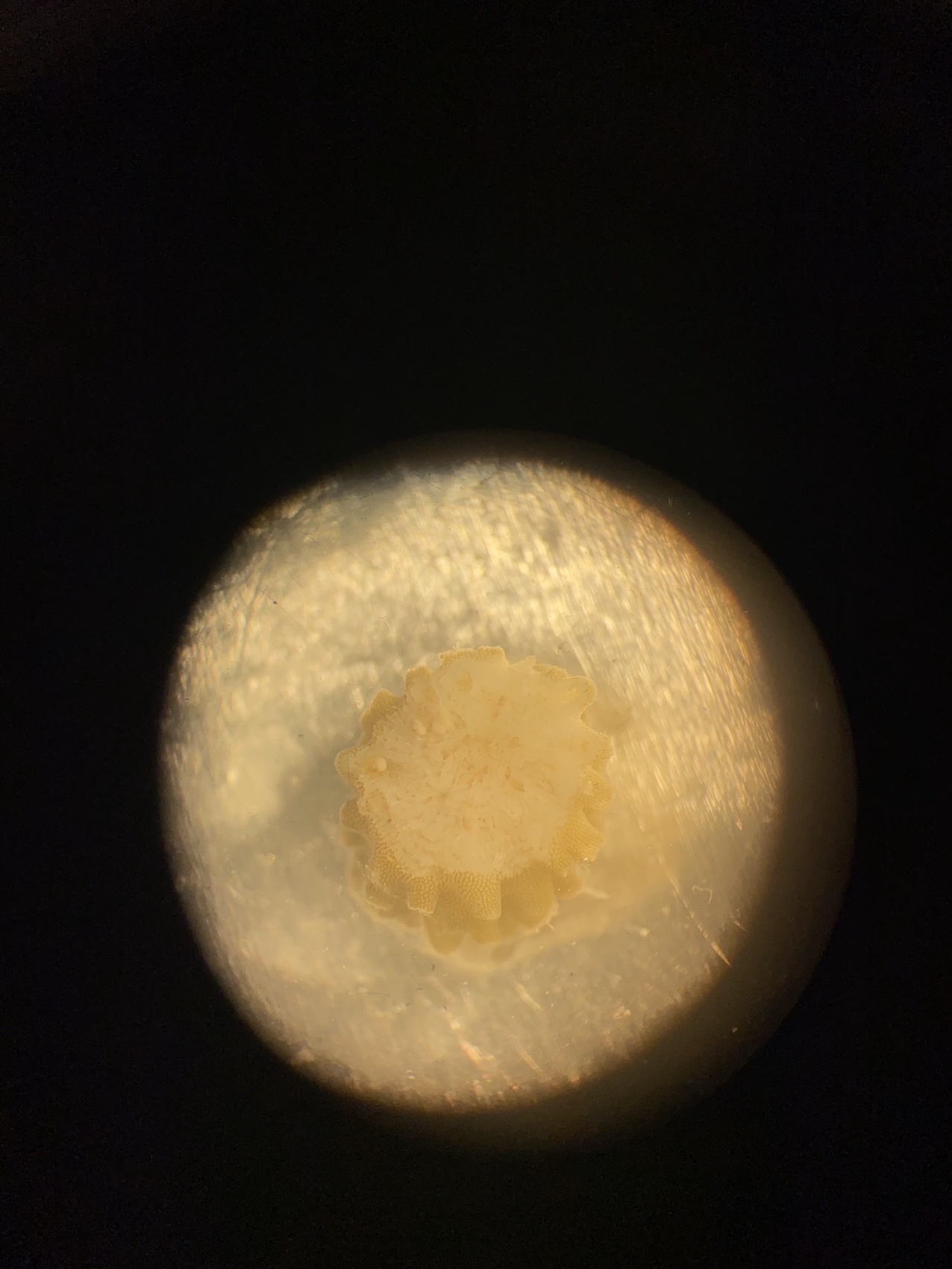
21 September 2022
Project set up
Ariana, Pierrick, and Danielle sourced materials for the E5 adult project including buying tank bins, finding shade cloth, and cleaning water tables. Light levels in the high light treatment will be 500-700 PAR peaking during the day and the low light treatment will reduce by 65% to approx. 100-300 PAR. See details on this project here.
Ariana also discussed experimental design with the E5 team and we have revised the experimental approach to include two sampling time points with fragments sourced from 10 colonies/genotypes.
Ariana will finish set up for this project this week and then move forward with fragging and starting the experiment once design is finalized.
A. pulchra high frequency sampling preparation
Danielle selected the bags and labeled tubes for the subset of coral colonies and tags that will be samples for the high frequency project. Bags and tubes for high frequency sampling will be labeled with HF colony ID (Ex: HF 416)
Metadata:
| Species | Site | OrderbyPatch | Original.Tag.Num | Patch | Latitude.S | Longitude.W | GPS.Point.Name | Timestamp | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. pulchra | Mahana | P1.3 | 414 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:59 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.5 | 416 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:11 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.4 | 424 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:17 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.1 | 426 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:49 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.1 | 429 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:05 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.4 | 430 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:10 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.9 | 431 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:46 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.5 | 433 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:02 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.4 | 434 | P1 | 17°29.234’ | 149°53.264’ | P1 | 1:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.10 | 436 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:29 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.2 | 437 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:50 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.10 | 439 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:47 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.1 | 448 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:35 PM | |||||||||||||||||
| A. pulchra | Mahana | P2.2 | 449 | P2 | 17°29.232’ | 149°53.253’ | P2 | 1:07 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.8 | 451 | P5 | 17°29.220’ | 149°53.236’ | P5 | 2:00 PM | |||||||||||||||||
| A. pulchra | Mahana | P5.6 | 458 | P5 | 17°29.220’ | 149°53.236’ | P5 | 1:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.11 | 461 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:30 PM | |||||||||||||||||
| A. pulchra | Mahana | P1.1 | 462 | P1 | 17°29.234’ | 149°53.264’ | P1 | 12:56 PM | |||||||||||||||||
| A. pulchra | Mahana | P4.6 | 464 | P4 | 17°29.231’ | 149°53.242’ | P4 | 1:41 PM | |||||||||||||||||
| A. pulchra | Mahana | P3.6 | 465 | P3 | 17°29.223’ | 149°53.249’ | P3 | 1:22 PM |
Boating
Danielle gave the team an orientation to driving the LTER boat (509) at Gump. Ariana took 4 trips as captain driving in the channels to learn navigation on the north shore. Ariana and Pierrick will work on obtaining their authorization for driving Gump boats.
20 September 2022
Project planning
Ariana developed a draft experimental design for the adult coral E5 repeat stressor experiment. A separate notebook post has been created here with all details and daily entries for this project in the E5 notebook. Ariana conducted planning and sourcing materials for this project today.
Overall, this project will expose coral colonies to ambient (450-600 PAR) and reduced (100-200 PAR) light conditions. A draft design is included below. This experiment will run until December. Monthly, fragments will be sampled for molecular, metabolomic, and physiological metrics. In December, S. Matsuda and R. Cunning will measure thermal phenotypes using the CBASS system.
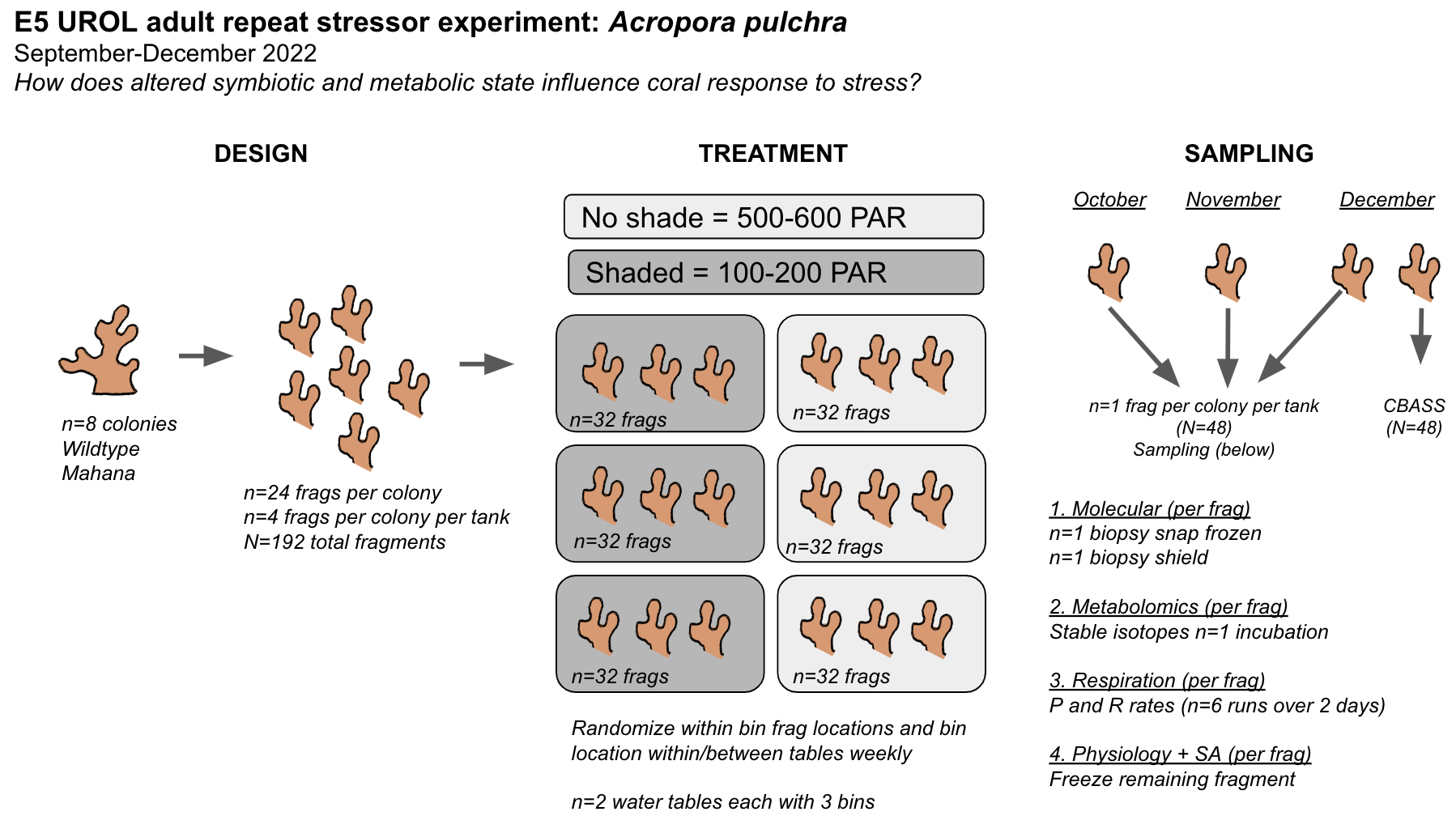
Tank cleaning
Ariana cleaned all tanks and larval conicals and stored for use in the next spawning event.
Physiological prep
Danielle sent all physiological protocols to Pierrick to prep for physiological processing of the September 2022 monthly sampling of A. pulchra starting this week. She also brought all the necessary equipment to the molecular lab for processing and got ziplock bags.
A. pulchra high frequency sampling planning
Danielle made a new folder for the A. pulchra high frequency histological sampling metadata and schedule for the next month before spawning. We will be processing a subset of our monthly timepoint A. pulchra samples for molecular (snap frozen and DNA/RNA shield) and histology (n=2 ~1 inch pieces of fragment preserved in 4% formalin solution) every five days to categorize and track gametogenesis very close to release. Danielle selected n = 4 from each of her five patches that had intact tags and will be sampling from ~0.5 m above them for the high frequency sampling period.
Freezer organization
Danielle moved all A. pulchra homogenate tubes to biocode -40 freezer and re-labeled a new shelf for Putnam lab. She updated all Mo’orea freezer/fridge lists with new samples. Organized all freezer boxes and freed up space for new samples in molecular freezers.
SDR calibration
We revised our approach to calibrating the SDR because we need to conduct calibrations at the level of individual wells. We can do this with a user-defined one point adjustment as specified in the SDR user manual and online resources.
The protocl is as follows and must be completed at the beginning of each measurement set (i.e., at the beginning of each day) at the temperature of measurement. If the plate will measure multiple temperatures, the one point adjustment will need to be conducted at that temperature. As long as the software remains open, multiple runs can be conducted with one set of calibrations.
Procedure:
- Aerate filtered seawater to generate 100% air saturated seawater (21% oxygen). Allow to fully aerate for 10-15 minutes.
- Set the incubators at the desired temperature.
- Load the SDR plate with the fully aerated seawater in each well ensuring there are no bubbles. Seal the wells with cover slips.
- Place the plates on SDRs in each incubator. Allow to come to temperature for 10 min.
- At this point, conduct the one point adjustment.
- Enter the correct measurement temperature and batch number in the software (PtS6-1624) and salinity.
- Select Calibration / One Point Adjustment
- The calibration will scan all wells.
- Enter pO2 (% Air Sat) = 100.
- Press OK
- The adjustment has been performed.
- Repeat at the start of each batch of measurements and at each respective temperature prior to measurement.
Loggers
Ariana downloaded all Hobo loggers from the larval tanks to test that loggers are recording data at correct intervals. Tomorrow we will conduct a calibration and offsets for all loggers.
19 September 2022
Project planning
Today we talked about project planning for the next few weeks in case the Acropora pulchra colonies do not spawn this month. If this is the case, spawning will occur at the following times:
October Full Moon = Oct 9 10:55am Tahiti time
A. pulchra: 9 nights after the full moon (Oct 18)
A. cytherea, A. retusa, A. hyacinthus: 6-9 nights after the full moon (Oct 15-18)
We are going to move forward with the following projects.
- Histological time series high frequency sampling
Danielle, Ariana, and Pierrick will conduct high frequency histology sampling of the A. pulchra histology time series of colonies at Mahana. We will collect samples from a subset of colonies (N=20 colonies, n=4 colonies per patch) every 5 days to characterize the final stages of gametogenesis in October.
- Processing histology time series physiology and histology samples
Danielle will train Pierrick how to process physiology and histology samples taken from the A. pulchra histology time series in September.
- E5 adult repeat stressor experiment
Ariana will plan and start an experiement under the E5 project to conduct a repeat stressor in adult corals. This project will expose fragments of collected wildtype A. pulchra to two light treatments. This will be conducted at Gump using bins that will either be covered with shade cloth or under natural light conditions.
The purpose of this project is to manipulate the metabolic condition of the coral holobiont and understand how metabolic changes influence stress response. A thermal stress test will be conducted in December with Ross Cunning and Shayle Matsuda using the CBASS system. This project will help us understand how metabolic condition influences stress response.
Throughout the adult exposure, we will characterize physiology, metabolism, and use stable isotope metabolomics to trace symbiotic nutritional exchange.
- October larval symbiont integration experiment
Ariana will conduct an experiment in october using the same experimental plan as described for September. Briefly, we will rear A. hyacinthus embryos to the larval stage, allow larvae to acquire symbionts, and expose the larvae to either ambient or elevated temperature conditions. Ariana will measure metabolism, survival, and nutritional exchange.
- Pocillopora population characterization
Pierrick will continue his project characterizing the haplotypes of Pocillopora spp. around Moorea. We will conduct additional sampling on transects at the Mahana and Hilton sites in September.
Spawning
We watched for spawning and no colonies spawned tonight. We will continue watching through Wednesday.
18 September 2022
Setting up temperature control
Ariana added heaters to the cooler systems in tanks A and C for high temperature control and connected to the Apex Neptune system. Temperatures programmed to be +4°C to see if the system can give us a high degree of elevated temperature without electrical limitations.



The script in the Apex program follows this temperature profile for each virtual stage.
29C - 30.4C (with 1.5C daily fluctuation)
Stage 1 = 29
Stage 2 = 29.2
Stage 3 = 29.4
Stage 4 = 29.6
Stage 5 = 29.8
Stage 6 = 30.0
Stage 7 = 30.2
Stage 8 = 30.4
This profile will be revised based on the diel fluctuation and the ambient temperature in Moorea. This profile is a test. The peak temperature occurs between 1500 and 1715 daily with this profile.
The script in the Apex system is as follows:
All master outlets
Set OFF
If Outlet High_Stage1 = ON Then ON
If Outlet High_Stage2 = ON Then ON
If Outlet High_Stage3 = ON Then ON
If Outlet High_Stage4 = ON Then ON
If Outlet High_Stage5 = ON Then ON
If Outlet High_Stage6 = ON Then ON
If Outlet High_Stage7 = ON Then ON
If Outlet High_Stage8 = ON Then ON
High_Stage1
Set OFF
If High_1 < 29 Then ON
If High_1 > 29 Then OFF
If Time 01:00 to 04:30 Then OFF
If Time 06:45 to 00:59 Then OFF
High_Stage2
Set OFF
If High_1 < 29.2 Then ON
If High_1 > 29.2 Then OFF
If Time 01:00 to 03:00 Then OFF
If Time 04:30 to 06:45 Then OFF
If Time 08:15 to 00:59 Then OFF
High_Stage3
Set OFF
If High_1 < 29.4 Then ON
If High_1 > 29.4 Then OFF
If Time 01:00 to 02:00 Then OFF
If Time 03:00 to 08:15 Then OFF
If Time 09:45 to 00:59 Then OFF
High_Stage4
Set OFF
If High_1 < 29.6 Then ON
If High_1 > 29.6 Then OFF
If Time 02:00 to 09:45 Then OFF
If Time 11:15 to 00:30 Then OFF
High_Stage5
Set OFF
If High_1 < 29.8 Then ON
If High_1 > 29.8 Then OFF
If Time 01:00 to 11:15 Then OFF
If Time 12:15 to 23:00 Then OFF
If Time 00:30 to 00:59 Then OFF
High_Stage6
Set OFF
If High_1 < 30 Then ON
If High_1 > 30 Then OFF
If Time 01:00 to 12:15 Then OFF
If Time 13:30 to 19:45 Then OFF
If Time 23:00 to 00:59 Then OFF
High_Stage7
Set OFF
If High_1 < 30.2 Then ON
If High_1 > 30.2 Then OFF
If Time 01:00 to 13:30 Then OFF
If Time 15:00 to 17:15 Then OFF
If Time 19:45 to 00:59 Then OFF
High_Stage8
Set OFF
If High_1 < 30.4 Then ON
If High_1 > 30.4 Then OFF
If Time 01:00 to 15:00 Then OFF
If Time 17:15 to 00:59 Then OFF
The temperature treatment was initiated at 1530 today. By 1800 the temperatures were at the desired level. We will let the profiles run for 24 h to view performance. Tomorrow we will look at the Apex data and download loggers.
Setting up SDR and lights
Ariana set up the incubators, SDR, and lights that will be required for respirometry and stable isotope incubations.

Aqua Illumination Prime 16HD reef lights are used to provide light in the incubators for photosynthesis measurements and stable isotope incubations.
Light settings were set to 40% intensity on all channels (UV, violet, royal, blue, green, deep red, cool white; no moonlight) positioned 25 cm above surface of the bottom of the incubator and 22 cm above the surface of the SDR plate. The goal is to obtain light at approx. 500 PAR at the level of the SDR plate.
Light was measured at 6 positions at the level of the SDR plate. This measurement should be conducted at the start of every day of measurement.
Incubators were labeled Incubator 1 and Incubator 2. Associated SDR 1 and 2 will be placed in the same number incubator.
Light measurements were recorded in Google Drive datasheet. Average light intensity was 494 PAR in Incubator 1 and 487 PAR in Incubator 2.
A HOBO Onset Pendant logger is placed in each incubator that can be launched at each respiration run to record temperature every 1 min.
Logger 21002976 is in Incubator 1 and logger 2133598 is in Incubator 2.


Calibrating SDR
We replaced oxygen sensor spots on the SDR microplates and need to calibrate since there are new spots from different batches than the old spots. This will also correct any drift of the spots.
SDR systems were assembled and set up and connected to the PreSens SDR software. To calibrate we followed the following steps:
- Mixed solution of 100% oxygen seawater by bubbling filtered seawater and made 0% oxygen seawater by adding sodium sulfite.
- Set oxygen unit to % O2 and record every 3 minutes. Select batch PSt5-1624.
- Select log measurement to create data for calibration labeled with Date and “calibration”. Saved on PC desktop in Moorea 2022 spawning folder. Will be uploaded to Google Drive and GitHub.
- Load half of wells with 0% solution and half with 100% oxygen solution in each plate. Record locations. Alternating wells with 0 and 100% water.
- Start measurements and let record for 2-3 h until stabilized recording every 3 min.
- Measure temperature with probe and loggers throughout.
- Export the data and calculate mean values for 0 and 100% wells from the last 15 min of measurement. We will create a new calibration dataset from these measurements and upload to the SDR software.
It is not clear yet how we perform a calibration for each well. We will look into this and try to do a well specific calibration this week.
Spawning
We watched for spawning and no colonies spawned tonight. We will continue watching through Wednesday.
17 September 2022
September 2022 Field Day Sampling - Backreef Acropora and Pocillopora
Danielle, Ariana, and Pierrick conducted monthly timepoint sampling at the Mahana field site (17°29’12.5”S 149°53’16.1”W). Following the Field Sampling Protocol, we collected 40 ~5 inch Acropora pulchra fragments from the five patches located on the site. Additionally, 10 4-5 inch fragments of Pocillopora were collected and brought back to Gump Station as well for same day processing. For all sampling locations, a close up photo with the tag was taken. Additionally, a bleaching score photo was taken at each sampling location. If a tag was not found, we sampled from the same colony that the tag should have been located on.
After collecting the samples, Danielle, Ariana, and Pierrick followed the Same Day Sample Processing Protocol to prepare samples for histological analysis, molecular and physiological downstream processing.
All bleaching score photos were downloaded off of the Putnam Lab Canon G7X camera number 2 and uploaded to Pocillopora_Sampling_Colony_Photos and Acropora_Sampling_Colony_Photos for the September 2022 timepoint.
All timepoint and tracking data sheets for the September 2022 timepoint for both Pocillopora and Acropora were updated.
Spawning
We again checked for spawning by stopping the water and pumps in all tanks at 2000 to watch for setting in collected colonies. We did not see setting by 2300. There was no spawning.
16 September 2022
Colony collection
Parent colonies were collected from the Mahana site on Moorea’s north shore (17°29’12.5”S 149°53’16.1”W).

We first collected n=24 A. pulchra experimental colonies for Danielle’s project at 0800-0900 at the Mahana field site. We transported these colonies back to Gump station at 1000. Parental colonies had previously experienced a marine heatwave scenario or ambient diel conditions in March-April 2022. All information on the heatwave experiment and data can be found here.
We then collected n=25 colonies (approx. 15-30cm in diameter) from large patches of Acropora pulchra from the Mahana field site between 1000-1200. We have a year-long project that has been tracking histology and physiology of colonies (Danielle’s project) at this site and therefore we have temperature and light data from this site as environmental context. The colonies collected are considered “wildtype” and were not taken from tagged colonies. Colonies were collected from different patches to obtain a genetically diverse sample.
Ariana and Danielle used a hammer and chisel to remove large fragments (15-30cm diameter) from these patches. Ariana Danielle and Pierrick transported them back to the boat where they were kept in coolers and buckets filled with seawater. Colonies were then transported back to Gump station at 1230 and placed in 2 flow through seawater tanks (n=12-13 colonies per tank). Tanks are equipped with 3 small recirculating pumps for water flow and had high water flow.

These colonies will be used to collect wildtype gametes to rear to larval stages for Ariana’s project. Spawn will be collected from each of the 2 tanks with wildtype colonies.
Launching loggers
Temperature loggers were launched to track temperature in the adult parent tanks (n=2 tanks for Danielle and n=2 tanks for Ariana; N=4 loggers) and larval conicals (n=4 conicals per blue tank; N=16 loggers).
Black Onset Hobo loggers were launched in each parent tank at 1900. Pendant Onset Hobo loggers were launched in larval conicals at 2200. Loggers are set to collect data every 10 minutes. At the end of the experiment we will need to do a group calibration to calculate offsets for the loggers. The loggers and location are as follows:
| Logger SSN | Tank |
|---|---|
| 20444040 | DBP Parent 2 |
| 20719656 | Wildtype Parent 2 |
| 20444033 | Wildtype Parent 1 |
| 20946644 | Wildtype Parent 1 |
| 21002982 | Conical A1 |
| 21002974 | Conical A2 |
| 21002979 | Conical A3 |
| 21002972 | Conical A4 |
| 21002981 | Conical B1 |
| 21335980 | Conical B2 |
| 21335987 | Conical B3 |
| 20937873 | Conical B4 |
| 21002977 | Conical C1 |
| 21335982 | Conical C2 |
| 20937872 | Conical C3 |
| 21002980 | Conical C4 |
| 21335986 | Conical D1 |
| 21335983 | Conical D2 |
| 21002975 | Conical D3 |
| 21335984 | Conical D4 |
Loggers were placed at the bottom of tanks. Pendants were positioned upright to measure light as well as temperature.
Apex system
Apex temperature probes were calibrated at 2230 using a digital thermometer (Traceable). Temperatures were between 26.1 and 26.4 for all tanks at the time of calibration.
Tank set up
We have been having problems with the water flow not keeping up with demand in the spawning systems with low flow coming from the cooler float valve. Ariana fixed this by cleaning the float valves to remove shell fragments and cutting tubing to go straight from the outlet to the float valve with no slack. To keep the system clean, we will purge the line before starting water flow to the coolers and will regularly check the float valves for shells or debris.
Tanks are now set up and running! We will add temperature control in the next few days.
Spawning
Spawning equipment and workflows were set up. Danielle prepared supplies and equipment to measure eggs per bundle, take samples of sperm and eggs for molecular responses, measure fecundity on sample fragments, and fertilize by parent treatment history. See Danielle’s notebook and the E5 public notebook for more information on her project.
Ariana gathered supplies needed for wildtype spawning. The protocol for Ariana’s spawning work will be as follows:
See a detailed post on this protocol here.
- Use gravy separators and mesh bottom cups to collect floating bundles from each of the large parent tanks. Count the number of parents that released to contribute to the gamete pool. Gather gametes into a bucket/bowl.
- Once all bundles are released, pool all gametes and gently mix.
- Alloquot 5 mL of bundles into each of 16-20 50 mL falcon tubes.
- Gently add 40 mL of filtered (50 um) seawater to each falcon tube.
- Allow falcon tubes to rest gently sideways with occassional gentle rocking for approx. 1 hour or until bundles are broken apart and eggs and sperm are allowed to mix.
- Remove the sperm water from the falcon tube using a pipette and add 10 mL of new filtered seawater to rinse the eggs. Remove this water.
- Pour the eggs into the conicals. Use 1 falcon tube for each conical. Have low water flow in the conicals.
- Place extra material if available into the 5 gallon buckets with filtered seawater using the same proceedure.
- Check the concial cultures every hour. Remove dead material with a pipette, gently mix, and use kim wipes to remove lipid films.
- Gently increase the water flow the next morning and continue hourly checks and cleaning.
We stopped the water and pumps in all tanks at 2000 to watch for setting in collected colonies. We did not see setting by 2300. There was no spawning. Red lights only were used to look at colonies.
15 September 2022
Set up
Today we continued setting up tanks and unpacking and organizing supplies. We continued assembling portable spawning systems in preparation for spawning. We made and purchased supplies for banjo filters for larval tanks and Gump staff assisted us with installing filtration systems for our water lines.
This is what our spawninig sytems look like and a full protocol is available here. These systems (pictured below) contain a cooler that acts as a header tank and can be controlled with heaters in combination with a Neptune Apex system. We will set this up in the comiing days. The water in the cooler is sourced from a seawater output. The water in the cooler is then pumped out to a PVC manifold that then delivers water to each larval tank. The larval tanks (i.e., conicals or “squaricals”) each have a banjo filter to allow for low thorugh seawater with large surface area to prevent embryos being damaged or lost on the drain. We set up four of these systems this week.



We set up 4 portable spawning systems each with a cooler and 6 larval conicals in a blue tank. Two of these tanks are for Ariana (labelled A and B) and two are for Danielle (labelled C and D). This provides each of us with a high and ambient temperature system. Tanks A and C will be the high temperature tanks and tanks B and D will be ambient temperature.
We also talked through plans for spawning and discussed potential protocols for measuring reproductive metrics for Danielle.
Finally, we prepared 6 extra buckets in a blue tank as extra space in case we have more gametes than can fit in the conicals. These buckets will contain seawater and sit in a water bath. They will not be flow through and will require water changes during embryo rearing.
Apex system
Ariana set up two units of Apex systems in preparation for temperature control in larval conicals. There is one Apex unit for Ariana and one unit for Danielle. This will allow for temperature monitoring in n=3 in each tank (A, B, C, D). The temperature of the conicals will be controlled based on the temperature readings in one high temperature conical. Each Apex system consists of the following parts:
1 Energy Bar AB8 1 Apex control unit 5 PM1 modules 6 Aquabus cables 6 Apex temperature probes
Apex units were connected to WiFi and created virtual outlets for each stage of temperature control. The Apex format looks like this:
We have the Apex systems named “AH Moorea Sq” and “DBP Moorea Sq”.
14 September 2022
Setup
Today we arrived in Moorea, French Polynesia. Goals for the day were to unpack supplies, assess space and materials and start experimental set up.
We will be constructing portable spawning systems developed in the Putnam Lab by Ariana Huffmyer and Jill Ashey. A full protocol for these systems and parts list is available here.
To prepare for spawning we cleaned and prepared tanks and started seawater flow. We will be using 4 large tanks to hold adult/parent colonies and 4 smaller tanks to hold the spawning systems. Ariana and Pierrick cleaned and filled tanks and constructed drain PVC systems for each tank. Danielle set up 24 twenty-two gallon tanks with designated labeling for the parental colonies that had previous experienced a diel marine heatwave, or ambient treatment in March-April 2022. Helped gather all supplies from around the station and organize lab space for spawning and subsequent sampling.

